Back to Journals » OncoTargets and Therapy » Volume 9
Clinicopathological significance of p15 promoter hypermethylation in multiple myeloma: a meta-analysis
Authors Wei B, Yang S, Zhang B, Feng Y
Received 17 December 2015
Accepted for publication 16 May 2016
Published 1 July 2016 Volume 2016:9 Pages 4015—4022
DOI https://doi.org/10.2147/OTT.S102733
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Li
Bing Wei, Shuhua Yang, Bo Zhang, Yong Feng
Department of Orthopaedic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China
Abstract: Published studies reported that loss of function of the p15INK4B gene is caused by hypermethylation; however, whether or not the inactivation is associated with the incidence and clinical significance of multiple myeloma (MM) remains unclear. In this study, we performed a meta-analysis to quantitatively determine the effects of p15 hypermethylation on the incidence of MM. The related research articles in English and Chinese languages were evaluated. The data were extracted and assessed independently. The pooled data were analyzed and odds ratios were calculated and summarized. Sixteen eligible studies were selected for final analysis. We demonstrated that p15 hypermethylation is significantly higher in MM than that in normal bone marrow, as well as monoclonal gammopathy of undetermined significance. However, aberrant p15 hypermethylation was not significantly higher in advanced MM than that in early-stage MM. The results of this study reveal that p15 hypermethylation is correlated with an increased risk in the progression of monoclonal gammopathy of undetermined significance to MM. p15 hypermethylation, which induces the loss of function of the p15 gene, plays a critical role in the early tumorigenesis of MM and serves as a reputable diagnostic marker and potential drug target.
Keywords: p15INK4B, methylation, asymptomatic monoclonal gammopathy of undetermined significance, tumor suppressor gene, odds ratio, meta-analysis
Introduction
Multiple myeloma (MM) is a clonal malignancy characterized by the production of monoclonal immunoglobulin and the proliferation of malignant plasma cells in the bone marrow.1 Clinically, MM starts with immortalization of a post-germinal center B cell and presents as asymptomatic monoclonal gammopathy of undetermined significance (MGUS). MGUS plasma cells share many abnormal characteristics with MM plasma cells. MGUS is considered as the precursor of MM, since it is able to progress to symptomatic MM at a rate of 1% per year.2 Over the years, newly approved drugs such as bortezomib, lenalidomide, and thalidomide have shown significant benefits in heavily pretreated MM patients.3–6 However, the majority of MM patients still relapse; therefore, the identification of biomarker and potential drug target is still necessary to improve the survival rate in the patients who are refractory to chemotherapy. Epigenetic alteration, particularly aberrant DNA methylation, is one of the best-characterized epigenetic modifications contributing to tumor initiation, progression, and prognosis.7–9 CpG islands of tumor suppressor genes are aberrantly methylated (hypermethylation), which result in transcriptional repression and loss of function of genes in many tumors including MM.10–12 The p15, as a tumor suppression gene, is hypermethylated in many tumors including MM.
p15INK4b (p15), one of the INK4 members, is a cyclin-dependent kinase inhibitor.13,14 Previously, p15INK4b, as well as other INK4 members, p14ARF, p16INK4a, p21CIP1, and p27KIP2, have been revealed to be involved in the neoplastic process of carcinomas.15,16 Although a number of reports have determined that the inactivation of p15 gene is mainly induced by hypermethylation in MM, the reported rates of p15 hypermethylation in MM are remarkably diverse. In addition, whether or not the inactivation of p15 gene is associated with the incidence and clinical significance of MM has not been thoroughly determined. Hence, we performed a systematic review and meta-analysis to determine the effects of p15 hypermethylation on the incidence and clinical significance of MM.
Methods
Search strategy and selection criteria
We identified studies from Embase, ISI web, and PubMed from August 1, 1995 to July, 2015 using the following search terms: “multiple myeloma”, “Kahler’s disease”, “plasma cell myeloma”, “methylation”, “p15”, “p15INK4B”, and “cyclin-dependent kinase inhibitor 2B”. We also manually searched reviews for additional related articles and the reference lists of the retrieved articles. We only took into account the studies published in the English and Chinese languages for full-text reading and final evaluations. After the exclusion of redundant and/or nonrelevant publications from different databases, the remaining articles were analyzed and evaluated in the full-text version for inclusion and exclusion criteria.
To be eligible, a study needed to meet the following criteria: 1) p15 methylation evaluated in primary MM; 2) p15 methylation determined by polymerase chain reaction (PCR); 3) research revealed the relationship between p15 methylation of MM clinicopathological parameters and prognosis; and 4) studies provided sufficient data and information to determine odds ratio (OR). The exclusion criteria included: 1) case reports, conference abstracts, letters, reviews, editorials, and expert opinion; and 2) all publications regarding in vitro/ex vivo studies, cell lines, and human xenografts. In addition to inclusion criteria, “aberrant” p15 methylation or p15 hypermethylation is defined by clear PCR product band detected by methylation-specific polymerase chain reaction (MSP).
Data extraction and methodological assessment
Two authors (BW, SY) independently reviewed and extracted data from eligible studies. Two authors (BZ, YF) reviewed all the articles that fit the inclusion and exclusion criteria. Disagreements were resolved through discussion and consensus. For each study, the following information was recorded: year of publication, the first author name, number of cases, sample source, methylation detection method, clinicopathological parameters, methylation rate, and follow-up results. Heterogeneity of investigations was evaluated to determine whether or not the data of the various studies could be analyzed.
Three investigators (BW, SY, and BZ) read through each publication independently for the methodological evaluation of the studies, and assessed and scored them according to the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines and European Lung Cancer Working Party (ELCWP) quality scale.17,18 They provided the quality scores and reached a consensus value for each item.
Statistical analysis
The Review Manager 5.2 (Cochrane Collaboration, Oxford, UK) and STATA 12.0 (StataCorp LP, College Station, TX, USA) were selected for analysis. The frequency of p15 promoter hypermethylation was compared in different tumor characteristics. The pooled frequency of p15 promoter hypermethylation and 95% confidence intervals (CIs) were estimated. Cochran’s Q test19 and the I2 statistic were used for evaluating heterogeneity among studies.20,21 A fixed-effect model was used to calculate parameters when heterogeneity was not an issue (I2 values <50%). A random-effects model was used to pool data and attempt to identify potential sources of heterogeneity based on subgroup analyses, if there was substantial heterogeneity (I2 values ≥50%). P-values tailed less than 0.05 were considered statistically significant.
The method reported by Egger et al was used for assessing publication bias.22 Meta-regression, subgroup analysis, and sensitivity analysis were used for exploring reasons of statistical heterogeneity. STATA version 10.0 was used for the analysis of meta-regression and publication bias.
Results
Sixteen studies published from 1997 to 2014 were included in the meta-analysis, as shown in Figure 1. A total of 607 patients from Argentina, Brazil, the People’s Republic of China, Hong Kong, France, Spain, Austria, and USA were enrolled; Table 1 showed their basic characteristics.
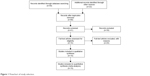  | Figure 1 Flowchart of study selection. |
The meta-analysis showed that P15 hypermethylation is significantly higher in MM than that in normal bone marrow samples. The pooled OR from 15 studies, including 571 MM and 193 normal bone marrow, are shown in Figure 2 (OR=19.80, 95% CI=9.21–42.56, P<0.00001), which indicates that p15 inactivation through hypermethylation plays an important role in the pathogenesis of MM. We further determined that p15 hypermethylation also occurs in MGUS, which is significantly less than that in MM (OR=2.25, 95% CI=1.20–4.21, P=0.01); the pooled frequency rate of the p15 hypermethylation in MM is 26.7% whereas in MGUS it is 13.9%, as shown in Figure 3A, but higher than that in normal bone marrow (OR=5.40, 95% CI=1.09–26.86, P=0.04), as shown in Figure 3B.
We analyzed 179 MM patients pooled in five studies to assess whether or not the aberrant p15 hypermethylation in MM was associated with advanced stages of MM. As shown in Figure 4, aberrant p15 hypermethylation is not significantly higher in advanced MM (Stage III) than that in early-stage MM (Stage I and II) (OR=1.17, 95% CI=0.54–2.54, P=0.69). Staging of MM is the process of finding out the amount of malignant myeloma cells that have advanced; it relies mainly on levels of albumin and beta-2-microglobulin in the blood, platelet count, kidney function, and patient’s age.23,24 MM stages which are equivalent to progression are determined by evolving mutations and epigenetic mechanisms. Our results suggest that the inactivation of p15 gene expression by promoter hypermethylation may not play an important role in MM progression.
Finally, we performed sensitivity analyses, in which one study at a time was removed, to assess the result stability. The pooled ORs were not significantly changed, indicating the stability of our analyses. The funnel plots are largely symmetric (Figure 5), suggesting there were no publication biases in the meta-analysis of p15 hypermethylation and clinicopathological features.
Discussion
The p15 gene, along with p14 and p16, is the most prevalent hypermethylated cell cycle regulatory gene in tumorigenesis and progression of MM.25–29 We conducted this meta-analysis to determine the correlation between p15 hypermethylation and MM. Analysis of the pooled data showed that 1) significantly higher p15 hypermethylation was detected in MM than that in normal bone marrow; 2) p15 hypermethylation was also detected in MGUS, but significantly less than that in MM; 3) p15 hypermethylation was also higher in MGUS than that in normal bone marrow; and 4) p15 hypermethylation was not significantly higher in advanced MM than that in early-stage MM. Epigenetic changes contribute to tumorigenesis and affect initial steps in malignant transformation by altering genome stability and regulating gene expression.30 MM develops as a result of multistep tumorigenic events, in which approximately one-third of all MM cases have a history of preceding MGUS or smoldering myeloma. Recent data also show that MM is consistently preceded by a precursor state, MGUS.31–33 MGUS, a premalignant and early stage of myeloma, was characterized by striking, widespread hypomethylation while gene-specific hypermethylation was found to occur in the advanced stages of myeloma.30 We noticed that Wang et al27 have performed a meta-analysis on p15 and p16 genes in MM; however, the role of p15 hypermethylation in different stages of MM was not elucidated. The results from the current study demonstrates that the hypermethylation rate of p15 gene promoter in MGUS was higher than that in normal bone marrow, indicating that p15 promoter hypermethylation was an early event in myelomagenesis. We did not find that p15 hypermethylation in MM was associated with advanced stage. These results also indicate that p15 hypermethylation could potentially be an early event. The results that significantly higher p15 hypermethylation was detected in MM than that in normal bone marrow and MGUS indicate that p15 hypermethylation is common in MM. Since changes in p15 hypermethylation are reversible, drug treatment through demethylation may be useful to delay carcinogenesis and progression. This approach may bring new direction and hope for MM treatment through gene-targeted therapy.
Progression from MGUS to MM appears to mirror the accumulation of genetic abnormalities, suggesting a stepwise progression of genetic and epigenetic changes.34 Hypermethylation of several tumor suppressors, such as p15, ARF, SOCS-1, p27 KIP1, RASSF1A, non-receptor type 6 (SHP1), death-associated protein kinase, and TP73 genes was reported in MGUS.35,36 Our meta-analysis showed that p15 gene hypermethylation in MGUS is higher than that in normal cohort. Our results also showed that p15 hypermethylation in MGUS was significantly less than that in MM. Therefore, hypermethylation of the p15 gene could potentially be involved in the progression of MGUS to MM. In addition, combining p15 with other tumor suppressor genes to develop several gene markers will be a valuable strategy for risk stratification to predict initial carcinogenesis and neoplastic progression of MM.
Limitations
This study has several potential limitations. First, the search strategy was restricted to articles published in English and Chinese. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. Second, the possibility of information and selection biases as well as unidentified confounders could not be completely excluded because all of the included studies were observational. Hence, caution should be taken when our findings are interpreted among the general populations. In addition, for MSP detection, it is usually to detect CpG island regions in the gene promoter or the first exon. The different location of detection sites may affect the methylation rate.
Conclusion
We performed a meta-analysis of 16 eligible studies. Our results demonstrated that p15 hypermethylation is significantly higher in MM than that in normal bone marrow, as well as in MGUS. However, aberrant p15 hypermethylation was not significantly higher in advanced MM than that in early-stage MM. The results of this meta-analysis suggest that p15 hypermethylation is associated with an increased risk in the progression of MGUS to MM. p15 hypermethylation, which induces the inactivation of p15 gene, plays an important role in the early tumorigenesis of MM and serves as a reputable prognostic marker and potential drug target.
Disclosure
The authors report no conflicts of interest in this work.
References
Rollig C, Illmer T. The efficacy of arsenic trioxide for the treatment of relapsed and refractory multiple myeloma: a systematic review. Cancer Treat Rev. 2009;35(5):425–430. | ||
Kyle RA, Therneau TM, Rajkumar SV, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med. 2002;346(8):564–569. | ||
Shank BR, Brown VT, Schwartz RN. Multiple myeloma maintenance therapy: a review of the pharmacologic treatment. J Oncol Pharm Pract. 2015;21(1):36–51. | ||
Latif T, Chauhan N, Khan R, Moran A, Usmani SZ. Thalidomide and its analogues in the treatment of multiple myeloma. Exp Hematol Oncol. 2012;1(1):27. | ||
Yang B, Yu RL, Chi XH, Lu XC. Lenalidomide treatment for multiple myeloma: systematic review and meta-analysis of randomized controlled trials. PLoS One. 2013;8(5):e64354. | ||
Romano A, Conticello C, Di Raimondo F. Bortezomib for the treatment of previously untreated multiple myeloma. Immunotherapy. 2013;5(4):327–352. | ||
Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029–15058. | ||
Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5(3):301–316. | ||
Fukushige S, Horii A. DNA methylation in cancer: a gene silencing mechanism and the clinical potential of its biomarkers. Tohoku J Exp Med. 2013;229(3):173–185. | ||
Chim CS, Kwong YL, Liang R. Gene hypermethylation in multiple myeloma: lessons from a cancer pathway approach. Clin Lymphoma Myeloma. 2008;8(6):331–339. | ||
Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5(5):553–568. | ||
Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem Biophys. 2013;67(2):501–513. | ||
Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995;9(15):1831–1845. | ||
Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56(4):722–727. | ||
Wolff L, Bies J. p15Ink4b Functions in determining hematopoietic cell fates: implications for its role as a tumor suppressor. Blood Cells Mol Dis. 2013;50(4):227–231. | ||
Heyman M, Einhorn S. Inactivation of the p15INK4B and p16INK4 genes in hematologic malignancies. Leuk Lymphoma. 1996;23(3–4):235–245. | ||
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97(16):1180–1184. | ||
Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18(4):705–719. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15(12):1237–1248; discussion 49–52. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Ribatti D, Moschetta M, Vacca A. Microenvironment and multiple myeloma spread. Thromb Res. 2014;133(Suppl 2):S102–S106. | ||
Ria R, Reale A, Vacca A. Novel agents and new therapeutic approaches for treatment of multiple myeloma. World J Methodol. 2014;4(2):73–90. | ||
Galm O, Wilop S, Reichelt J, et al. DNA methylation changes in multiple myeloma. Leukemia. 2004;18(10):1687–1692. | ||
Guillerm G, Depil S, Wolowiec D, Quesnel B. Different prognostic values of p15(INK4b) and p16(INK4a) gene methylations in multiple myeloma. Haematologica. 2003;88(4):476–478. | ||
Wang X, Zhu YB, Cui HP, Yu TT. Aberrant promoter methylation of p15 (INK4b) and p16 (INK4a) genes may contribute to the pathogenesis of multiple myeloma: a meta-analysis. Tumour Biol. 2014;35(9):9035–9043. | ||
Braggio E, Maiolino A, Gouveia ME, et al. Methylation status of nine tumor suppressor genes in multiple myeloma. Int J Hematol. 2010;91(1):87–96. | ||
Ng MH, Chung YF, Lo KW, Wickham NW, Lee JC, Huang DP. Frequent hypermethylation of p16 and p15 genes in multiple myeloma. Blood. 1997;89(7):2500–2506. | ||
Heuck CJ, Mehta J, Bhagat T, et al. Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis. J Immunol. 2013;190(6):2966–2975. | ||
Landgren O. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: biological insights and early treatment strategies. Hematology Am Soc Hematol Educ Program. 2013;2013:478–487. | ||
Agarwal A, Ghobrial IM. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: a review of the current understanding of epidemiology, biology, risk stratification, and management of myeloma precursor disease. Clin Cancer Res. 2013;19(5):985–994. | ||
Greenberg AJ, Rajkumar SV, Vachon CM. Familial monoclonal gammopathy of undetermined significance and multiple myeloma: epidemiology, risk factors, and biological characteristics. Blood. 2012;119(23):5359–5366. | ||
Iida S, Ueda R. Multistep tumorigenesis of multiple myeloma: its molecular delineation. Int J Hematol. 2003;77(3):207–212. | ||
Stanganelli C, Arbelbide J, Fantl DB, Corrado C, Slavutsky I. DNA methylation analysis of tumor suppressor genes in monoclonal gammopathy of undetermined significance. Ann Hematol. 2010;89(2):191–199. | ||
Chim CS, Liang R, Leung MH, Kwong YL. Aberrant gene methylation implicated in the progression of monoclonal gammopathy of undetermined significance to multiple myeloma. J Clin Pathol. 2007;60(1):104–106. | ||
Li C, Yang L, Liu L, et al. Study on methylation and mRNA expression of p15 gene in multiple myeloma. Med Pharm Yunnan. 2014;3:1–3. Chinese. | ||
Martin P, Garcia-Cosio M, Santon A, Bellas C. Aberrant gene promoter methylation in plasma cell dyscrasias. Exp Mol Pathol. 2008;84(3):256–261. | ||
Jiang J, Lin S, Chen G, Yan J. The significance of p15 gene methylation in multiple myeloma. J Clin Hematol. 2007;20(2):104–105. Chinese. | ||
Liang X, Jiang Y, Xu W, Liu J. Study on hypermethylation and mRNA expression of p16 and p15 gene in multiple myeloma. Clin Focus. 2006;21(1):10–13. | ||
Chim CS, Kwong YL, Fung TK, Liang R. Methylation profiling in multiple myeloma. Leuk Res. 2004;28(4):379–385. | ||
Seidl S, Ackermann J, Kaufmann H, et al. DNA-methylation analysis identifies the E-cadherin gene as a potential marker of disease progression in patients with monoclonal gammopathies. Cancer. 2004;100(12):2598–2606. | ||
Chen W, Wu Y, Zhu J, Liu J, Tan S, Xia C. Methylation of p16 and p15 genes in multiple myeloma. Chin Med Sci J. 2002;17(2):101–105. | ||
Fu W, Gao W, Hou J, Wang D, Ding S, Chen Q. Hypermethylation of CpG island of p15 and p16 genes in multiple myeloma. Cancer Prev Cure Res. 2002;2(29):1–2. | ||
Guo X, Fan H, Tan G, et al. Study of p15 gene methylation status in multiple myeloma. Chin J Pathophysiol. 2001;17(7):679–682. | ||
Guillerm G, Gyan E, Wolowiec D, et al. p16(INK4a) and p15(INK4b) gene methylations in plasma cells from monoclonal gammopathy of undetermined significance. Blood. 2001;98(1):244–246. | ||
Wu Q, Guo X, Fan H, et al. [P15(INK4B) gene methylation in malignant hematopoietic diseases]. Zhonghua Xue Ye Xue Za Zhi. 2000;21(12):644–646. Chinese. | ||
Fan H, Guo X, Wu Q, et al. [Detection of high methylation of p15(INK4B) and p16(INK4A) genes in multiple myeloma]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2000;8(4):271–274. Chinese. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





