Back to Journals » OncoTargets and Therapy » Volume 9
Clinicopathological role of miR-30a-5p in hepatocellular carcinoma tissues and prediction of its function with bioinformatics analysis
Authors Huang W, Chen Z, He R, Wu Y, Yin S, Liang X, Chen G , Yang H, Peng Z , Yang L
Received 26 April 2016
Accepted for publication 19 May 2016
Published 12 August 2016 Volume 2016:9 Pages 5061—5071
DOI https://doi.org/10.2147/OTT.S111431
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Li
Wen-Ting Huang,1,* Zu-xuan Chen,2,* Rong-quan He,2 Yu-zhuang Wu,1 Shu-ya Yin,1 Xiao-na Liang,1 Gang Chen,1 Hong Yang,3 Zhi-gang Peng,2,* Li-hua Yang2,*
1Department of Pathology, 2Department of Medical Oncology, 3Department of Ultrasonography, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China
*These authors contributed equally to this work
Background: It has been reported that deregulation or dysfunction of microRNAs (miRNAs) plays an essential part in the hepatocarcinogenesis. However, the contribution and mechanism of microRNA-30a-5p (miR-30a-5p) in hepatocellular carcinoma (HCC) remains largely unknown. Therefore, our aim was to investigate the clinicopathological role of miR-30a-5p in HCC tissues and explore its potential pathways in this study.
Methods: The expression of miR-30a-5p was measured in 95 HCC and adjacent noncancer tissues by real-time reverse transcription quantitative polymerase chain reaction. The relationship between miR-30a-5p expression levels and clinicopathological parameters was also analyzed. Furthermore, the potential target genes of miR-30a-5p were collected via online prediction and literature searching. Gene ontology and pathway enrichment analyses were used to identify the possible function of miR-30a-5p in HCC.
Results: Compared with adjacent noncancer tissues (2.23±0.77), expression level of miR-30a-5p was significantly lower in HCC tissues (1.26±0.66, P<0.001). MiR-30a-5p expression was evidently correlated with tumor nodes, metastasis, tumor–node–metastasis stage, portal vein tumor embolus, vascular invasion, and status of tumor capsule (all P<0.05). A total of 878 genes were finally used for the biological informatics analyses. These prospective target genes were highly enriched in various key pathways, for instance, Ubiquitin-mediated proteolysis, Axon guidance, Neurotrophin signaling pathway, Amyotrophic lateral sclerosis, and ErbB signaling pathway.
Conclusion: In conclusion, this study clarifies that the downregulation of miRNA-30a-5p might play a vital part in the incidence and progression of HCC via targeting various prospective genes and pathways. Future validation is required to further explore the prospective molecular mechanism of miR-30a-5p in HCC.
Keywords: miR-30a-5p, hepatocellular carcinoma, progression, target genes, gene ontology analysis, pathway analysis
Introduction
Liver cancer is one of the most frequent cancers worldwide with a progressively increasing tendency of mortality and morbidity worldwide. According to statistical data, 28,410 new estimated cases of liver and intrahepatic bile duct cancer will occur in males in 2016, which ranks tenth and accounts for 3% of all malignancies. More importantly, liver and intrahepatic bile duct cancer is the leading cause of death in both males (estimated deaths 18,280, ranking fifth) and females (estimated deaths 8,890, ranking eighth, 3%) in the United States.1 On a more serious note, in People’s Republic of China liver cancers were the most commonly diagnosed cancer and the most prevalent cause of cancer-related death, after lung, stomach, and esophageal cancers (based on the statistics in 2015).2 In liver cancer, hepatocellular carcinoma (HCC) is the most common class, which comprises about 90% of cases.3 At the time of definitive diagnosis of HCC, the disease has frequently reached an advanced stage and curative treatment opportunities are often lost; thus, the recurrence rates of HCC still remain high.4 Survival for HCC patients after diagnosis is not optimistic, with a median survival time about 6–20 months.5 Furthermore, HCC is unevenly distributed in the world based on the different leading risk factors.3 Genetic markers, including AFP, cannot be used due to the heterogeneity of HCC. Therefore, one of the more difficult tasks in clinical research of HCC is to attempt to discover novel markers to improve the diagnosis technique and to boost the survival prediction and treatment indication for HCC patients.5,6
MicroRNA (miRNA) is a class of noncoding RNAs that contain 19–25 nucleotides.7 These miRNAs can target the 3′-untranslated region of mRNA, which subsequently leads to mRNA degradation or translational repression. miRNAs are widely involved in the biological functions such as cell proliferation, differentiation, and apoptosis.8 On the basis of studies, it has been estimated that more than 60% human genes can be regulated by different miRNAs, and synchronously, one single miRNA can target various mRNAs to fulfill its biological functions.9 Growing evidence has revealed that miRNAs play a vital role in HCC.10–13 Previously, we found that overexpression of microRNA-30a-5p (miR-30a-5p; accession number: MIMAT0000087) in vitro could markedly inhibit cell growth and induce caspase-3/7 activity and apoptosis in four HCC cell lines HepG2, SMMC-7221, HepB3, and SNU449,14 consistent with the in vitro finding by Liu et al15 and Li et al.16 They also detected the expression of miR-30a-5p in HCC samples; however, the number of patients involved was extremely small (n=63 in Liu et al15 and n=16 in Li et al16). The clinicopathological significance of miR-30a-5p in HCC remains largely unclear. Thus, we were interested in analyzing the expression status of miR-30a-5p in HCC tissues by real-time reverse transcript-quantitative polymerase chain reaction (RT-qPCR), and also studying the correlations between miR-30a-5p expression and clinicopathological parameters of HCC. Further, we performed bioinformatics analysis to gather the possible target genes and potential pathways of miR-30a-5p in regulation gene network of HCC.
Materials and methods
Tissue samples and RT-qPCR
A total of 95 patients from the First Affiliated Hospital of Guangxi Medical University, People’s Republic of China (between March 2010 and December 2011) were included in this study. The age of HCC patients ranged from 29 to 82 years, with a mean age of 52 years. The detailed clinicopathological information of patients is summarized in Table 1. Tissue samples were obtained from surgical resection of patients who had not received any treatment. The adjacent noncancer hepatic tissues were taken at least 2 cm away from the border of the tumor observed by naked eyes and confirmed independently by two pathologists (Wen-ting Huang and Gang Chen) as being without cancer by microscopic analysis. This study was approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University, and written informed consent was obtained from each participant, according to the institutional guidelines of our hospital.
The expression of miR-30a-5p was detected by real-time RT-qPCR. First, total RNA (including miRNA) was extracted from all samples with the miRNeasy FFPE Kit (QIAGEN, the Netherlands), following the manufacturer’s protocol as previously reported.17–23 RNA concentrations were determined using the Nano Drop 2000. A combination of RUN6B and RUN48 was the housekeeping reference used in this study. The primers for miR-30a-5p, RNU6B, and RNU48 were included in TaqMan® MicroRNA Assays (4427975, Applied Biosystems, Life Technologies, Grand Island, NY, USA). Primer sequences were as follows: miR-30a-5p (Applied Biosystems, cat no 4427975-000417): UGUAAACAUCCUCGACUGGAAG; RNU6B (Applied Biosystems, cat no 4427975-001093): CGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU; RNU48 (Applied Biosystems, cat no 442975-001006): GAUGACCCCAGGUAACUCUGAGUGUGUCGCU GAUGCCAUCACCGCAGCGCUCUGACC. The reverse primers were also included in the process of reverse transcription with TaqMan®MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems) to a total volume of 10 μL. Real-time qPCR to detect the miRNA level was performed using the Applied Biosystems PCR7900. The miR-30a-5p level in each sample was normalized to its internal references. The expression of miR-30a-5p level in the FFPE experiments was calculated with the formula 2−Δcq as previously reported.17–23
Collection of prospective target genes of miR-30a-5p via bioinformatics approaches
To obtain a better picture of the potential mechanism of miR-30a-5p in the progression of HCC, the validated target genes of miR-30a-5p from literature screening were collected from PubMed, Wiley Online Library, Web of Science, Science Direct, Cochrane Central Register of Controlled Trials, Google Scholar, EMBASE, Ovid, and LILACS. Since the previous name of miR-30a-5p was miR-30a and there are several synonyms of miR-30a-5p available, the following searching key words were applied: “(miR-30a or miRNA-30a or microRNA-30a or miR30a or miRNA30a or microRNA30a or ‘miR 30a’ or ‘miRNA 30a’ or ‘microRNA30a’ or miR-30a-5p or miRNA-30a-5p OR microRNA-30a-5p) and (target*).” Concurrently, target genes with “strong evidence” were also gathered from mirTarBase and TarBase databases. All the target genes from aforementioned sources verified by qPCR, Western blot, or luciferase reporter assay were integrated into “validated targets” of miR-30a-5p. To further acquire a more complete image of potential target genes of miR-30a-5p, we predicted the possible targets via 11 online methods, including MirTarBase, TarBase, Targetminer, polymiRTS, RNA22, microRNA.org, Pita, mirRNAMAP, Targetscan, miRDB, and Pictar-vert. Genes appearing more than five times were selected as the “predicted targets” of miR-30a-5p.
Bioinformatics analyses with gene ontology (GO) and pathway enrichment
The genes merged from “validated targets” and “predicted targets” were evaluated for further GO and pathway analysis with the DAVID (https://david.ncifcrf.gov/) and BINGO plug-in of Cytoscape. Genes were also mapped to the database of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway to identify the signaling. P-value <0.001 was of significance in GO analysis. Similarly, P<0.001 was considered significant in pathway analysis.
Statistical analysis
All statistical analyses were conducted using SPSS version 20.0 (Armonk, NY, USA). The expression levels were presented as mean ± standard deviation. Receiver operator characteristic curve (ROC) was applied to evaluate the diagnostic value. P-value <0.05 was considered of statistical significance.
Results
Clinicopathological significance of miRNA-30a-5p in HCC tissues
In the studied population of HCC patients, the expression of miRNA-30a-5p was significantly downregulated in HCC tissues (1.2564±0.6561) compared with that in adjacent noncancer hepatic tissues (2.2309±0.7677, P<0.001, Figure 1). The area under the ROC curve of low miRNA-30a-5p was 0.857 (95% CI: 0.801–0.912; P<0.001; cut-off =1.395), with 87.4% sensitivity and 18.9% specificity in distinguishing the HCC from noncancerous liver tissues (Figure 2). The level of miRNA-30a-5p was also reduced in patients with tumors in multiple nodes, metastasis, tumor–node–metastasis (TNM) stage III or IV disease, portal vein tumor thrombus, vascular invasion, tumor capsule infiltration or no capsule, and low microvessel density, when compared with the counterparts (Table 1). The association of miRNA-30a-5p with clinicopathological parameters was further supported by Spearman analysis. From the results, miRNA-30a-5p was shown to be negatively associated with TNM stages (r=−0.213; P=0.038), metastasis (r=−0.516; P<0.001), tumor nodes (r=−0.317; P=0.002), tumor capsule (r=−0.255; P=0.013), portal vein tumor thrombus (r=−0.615; P<0.001), and vascular invasion (r=−0.407; P<0.001). We also investigated the relationship between miR-30a-5p and recurrence. Seventy patients in this study were followed up and all of them had recurrent disease. The follow-up time of HCC patients range from 2.68 to 68 months. The mean recurrent time was 32.30±1.76 months. We divided these 70 patients with follow-up information into two groups based on the cut-off of 0.98. There was no significant difference of the recurrent time between patients having high miR-30a-5p level (32.29±2.4) and low level (33.25±2.78, P=0.533; Figure 3).
  | Figure 1 MiR-30a-5p expression in HCC and adjacent noncancer liver tissues. |
  | Figure 2 ROC curve of miR-30a-5p expression in HCC. |
  | Figure 3 Relationship between miR-30a-5p expression and recurrence of HCC. |
Bioinformatics analysis of the potential target genes of miR-30a-5p
The confirmed target genes (n=64) of miR-30a-5p in the literature were recorded. In addition, another 30 genes with strong evidence, which were validated by qPCR, Western blot, or luciferase assays, were provided by mirTarBase and TarBase databases. Seventeen genes were present at the same time in the literature and in the mirTarBase/TarBase database (RUNX2, VIM, DTL, SEPT7, MTDH, Snai1, PRDM1, AVEN, FOXD1, BDNF, BECN1, TNRC6A, CDH1, BCL11A, HSPA5, EYA2, and SOX4). Thus, the disjoint union of 77 genes were considered as “validated targets” of miR-30a-5p. Afterward, 11 online software predicted 9,133 target genes. To decrease the false positivity of target genes, 829 genes that came up more than five times were grouped into “predicted targets” of miR-30a-5p. After “validated targets” and “predicted targets” were combined, we finally had 878 genes for the following bioinformatics analyses (Figure 4).
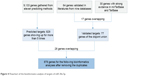  | Figure 4 Flowchart of the bioinformatics analysis of targets of miR-30a-5p. |
According to the results of DAVID and BINGO in GO analysis, the target genes were found concentrated in the following biological pathways: regulation of transcription from RNA polymerase II promoter, positive regulation of cellular biosynthetic process, and positive regulation of biosynthetic process (P<0.001, Figure 5). On the basis of cellular component, genes mostly assembled at the pathways of Golgi apparatus, insoluble fraction and membrane fractions, etc (P<0.001, Figure 6). Genes prominently accumulated in four molecular functions, including transcription regulator activity, protein domain-specific binding, transcription activator activity, and acid-amino acid ligase activity (P<0.001, Figure 7). Besides, KGEE pathway analysis showed that five pathways were significant, ie, Ubiquitin-mediated proteolysis, Axon guidance, Neurotrophin signaling pathway, Amyotrophic lateral sclerosis, and ErbB signaling pathway (P<0.05, Figure 8).
  | Figure 8 Pathways of the prospective target genes of miR-30a-5p by KEGG analysis. |
Discussion
Previously, we found that miR-30a-5p expression was lower in the HCC cell lines compared to the normal hepatic cells (data not shown), which was in agreement with the studies of Liu et al15 and Li et al.16 Thus far, only two groups have attempted to investigate the difference of miR-30a-5p level between HCC tissues and noncancerous liver tissues. Li et al16 used RT-qPCR to detect miR-30a-5p expression in 16 pairs of HCC and their adjacent noncancerous tissues. Among 16 cases, 13 (81.25%) presented lower expression of miR-30a-5p in HCC tissues compared with matched noncancerous liver tissues. The results showed that miR-30a-5p expression in HCC tissues was significantly lower compared to adjacent noncancerous liver tissues. Besides, Liu et al15 collected human HCC tissues and their paracancerous hepatic tissues from 63 patients undergoing resection and showed that miR-30a was downregulated in 87% (55/63) of the examined HCC tissues. In accordance with the aforementioned two reports, we also confirmed the striking downregulation of miR-30a-5p expression in HCC tissues, compared to the noncancerous liver controls, with a larger cohort of 95 pairs of clinical samples. The expression of miR-30-5p was only 56% that in the noncancerous liver, which indicates that during the process of HCC development, miR-30a-5p is lost. Hence, miR-30a-5p probably plays a tumor suppressive role in the carcinogenesis of HCC. Furthermore, ROC revealed that in this study, the diagnostic value of low miR-30a-5p level remained moderate with an area under the curve of 0.875. MiR-30a-5p might also act as a biomarker contributing in the screening of HCC. However, a larger study is warranted for confirmation of the diagnostic value of miR-30a-5p for HCC in the future.
miRNAs are particularly stable in body fluids, including serum or plasma, which makes them preferable biomarkers for the early diagnosis in HCC.24,25 To date, only one study has identified and evaluated miR-30a-5p as a HCC-associated plasma miRNA in HCC. The result showed that miR-30a-5p was one of the significantly overexpressed miRNAs in the hepatitis B virus-positive HCC patients compared with the hepatitis B virus-positive cancer-free controls,26 which is a contrary phenomenon compared to its reduced expression level in HCC tissues. The cause of this contradiction needs further investigation.
With regard to the correlation between miR-30a-5p and clinical parameters of HCC, Liu et al’s15 study was the only one that explored whether miR-30a-5p downregulation was associated with clinical features or prognosis of HCC patients. A relationship between reduced miR-30a expression and intrahepatic metastasis, advanced TNM stage, and high Edmonson pathological classification was noted. Additionally, lower miR-30a level was associated with shorter disease-free survival, and multivariate analysis confirmed that low miR-30a-5p level was an independent predictor for shorter disease-free survival of HCC patients (HR =3.2; P=0.002). In this study, we found a similar relationship between low miR-30a-5p and the deterioration of HCC, which is associated with the status of metastasis, number of tumor nodes, condition of tumor capsule, portal vein tumor thrombus, and vascular invasion, although no significant correlation of miR-30a-5p was noted with recurrence. Collectively, the data of this study, together with the finding of Liu et al,15 suggest that downregulation of miR-30a-5p may play an important part in the development and progression of HCC.
The clinicopathological significance of miR-30a-5p in HCC has also been verified with in vitro experiments. Previously, our group performed in vitro work to investigate the role of miR-30a-5p on the biological function of HCC cell lines HepG2, SMMC-7221, HepB3, and SNU449. We transfected miR-30a-5p inhibitor and miR-30a-5p mimic into HCC cell lines and found that miR-30a-5p mimic could obviously inhibit cell growth and also induce caspase-3/7 activity and apoptosis.14 Li et al16 also reported similar results that miR-30a-5p overexpression in HCC cells significantly inhibited cell proliferation, suppressed colony-forming efficiency, induced apoptosis in vitro, and also reduced HepG2 tumor growth in vivo. Furthermore, gain- and loss-of-function studies by Liu et al15 demonstrated that downregulation of miR-30a-5p facilitated tumor cell migration, invasion, and epithelial–mesenchymal transition. In summary, miR-30a-5p could influence several malignant phenotypes including cell growth, migration, invasion, apoptosis, and epithelial–mesenchymal transition of HCC cells.
Next, the molecular mechanism of miR-30a-5p is of great interest. Almost at the same time, Li et al16 and our group both reported that metadherin (MTDH)/AEG-1 was one of the direct target genes of miR-30a-5p, which was confirmed by luciferase assay. Besides, Liu et al15 identified SNAI1 also as a direct target of miR-30a-5p. These two genes, MTDH/AEG-1 and SNAI1, are by far the only validated targets of miR-30a-5p in HCC. Since one single miRNA can target dozens of genes to fulfill its biological and clinical function, the relevant gene network and molecular mechanism of miR-30a-5p remain undefined. Thus, the bioinformatics analysis was performed to comprehensively understand the prospective targets and pathways of miR-30a-5p. Various pathways have been shown with GO and KEGG analyses, which suggests that it could be a complicated process for miRNA-30a-5p to regulate different signaling pathways. This is also accordance with the accepted idea that HCC is a cancer, implying the multistep process of activating oncogenes and inactivating tumor suppressor genes. As the strong connection of miRNA-30a-5p expression and tumor progression was noted in this study, we paid more attention to the pathways related to cell grow, cell invasion, and metastasis. Among all the significant pathways shown by GO and KEGG analyses, some have been widely studied in HCC. For example, PIK3CD, TP53, FOXO3, IRS1, AKT1, MAPK1 in both Neurotrophin signaling pathway and ErbB signaling pathway have been confirmed to play vital roles in HCC.27–34 Since these potential targets genes of miR-30a-5p have never been confirmed to date, future verification studies are expected to verify more target genes and related pathways of miR-30a-5p in HCC.
Autophagy has been considered as a prospective target to enhance the efficiency of conventional chemotherapeutics for HCC.35,36 MiR-30a-5p, a powerful inhibitor of autophagy by restraining Beclin-1, was found to be interfering with the effectiveness of sorafenib-mediated apoptosis by an autophagy-dependent pathway in renal cell carcinoma cells.37 Sorafenib is by far the only and standard systematic therapy drug for the treatment of advanced HCC, and its clinical benefits remain modest. Little is known of the biomarker to predict the efficiency of sorafenib on HCC, or the molecular mechanism of the drug resistance.38,39 MiR-30a-5p might be a hopeful link to connect sorafenib-induced autophagy activation and HCC resistance.
It should be emphasized that limitation exists in this study. First, the survival data are missing due to the fact that most patients were lost after they were discharged from our hospital. The prognostic value of miR-30a-5p in HCC needs further verification. Second, even though thousands of prospective genes have been predicted in this study, only two genes (MTDH/AEG-1 and SNAI1) have been experimentally validated. More target genes are expected to be verified in the future work. Third, the retrospective nature of this study is itself a limitation. Prospective studies with a larger sample size are needed for further confirmation of the role of miR-30a-5p in HCC. Moreover, our research only aimed at HCC patients of Chinese yellow race. Since racial variations might have impact on the molecular signature of HCC, studies with a larger sample size and including other races are required in the future.
Conclusion
This study clarifies that the downregulation of miRNA-30a-5p might play a vital role in the incidence and progression of HCC via targeting various prospective genes and pathways. Future validation is required to further explore the molecular mechanism of miR-30a-5p in HCC.
Acknowledgments
This study was supported by the fund of Future Academic Star of Guangxi Medical University (WLXSZX16042), Guangxi Zhuang Autonomous Region University Student Innovative Plan (201510598016), Guangxi Natural Science Foundation (2015GXNSFBA139157), and National Natural Science Foundation of China (NSFC81260222).
Disclosure
The authors report no conflicts of interests in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. | ||
Plentz RR, Malek NP. Early detection of hepatocellular carcinoma. How to screen and follow up patients with liver cirrhosis according to the GERMAN S3 guideline? Diagnostics. 2015;5(4):497–503. | ||
Saraswat VA, Pandey G, Shetty S. Treatment algorithms for managing hepatocellular carcinoma. J Clin Exp Hepatol. 2014;4(Suppl 3):S80–S89. | ||
Diaz-Gonzalez A, Forner A, Rodriguez de Lope C, Varela M. New challenges in clinical research on hepatocellular carcinoma. Rev Esp Enferm Dig. Epub 2015 Dec 10. | ||
Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21(34):9853–9862. | ||
Arabkheradmand A, Safari A, Seifoleslami M, Yahaghi E, Gity M. Down-regulated microRNA-124 expression as predictive biomarker and its prognostic significance with clinicopathological features in breast cancer patients. Diagn Pathol. 2015;10(1):178. | ||
Pan HL, Wen ZS, Huang YC, et al. Down-regulation of microRNA-144 in air pollution-related lung cancer. Sci Rep. 2015;5:14331. | ||
Wu SY, Lan SH, Liu HS. Autophagy and microRNA in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2016;22(1):176–187. | ||
Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21(37):10584–10597. | ||
Mao B, Wang G. MicroRNAs involved with hepatocellular carcinoma (Review). Oncol Rep. 2015;34(6):2811–2820. | ||
Lyra-Gonzalez I, Flores-Fong LE, Gonzalez-Garcia I, Medina-Preciado D, Armendariz-Borunda J. MicroRNAs dysregulation in hepatocellular carcinoma: Insights in genomic medicine. World J Hepatol. 2015;7(11):1530–1540. | ||
He R, Yang L, Lin X, et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8(12):15632–15641. | ||
Liu Z, Tu K, Liu Q. Effects of microRNA-30a on migration, invasion and prognosis of hepatocellular carcinoma. FEBS Lett. 2014;588(17):3089–3097. | ||
Li WF, Dai H, Ou Q, Zuo GQ, Liu CA. Overexpression of microRNA-30a-5p inhibits liver cancer cell proliferation and induces apoptosis by targeting MTDH/PTEN/AKT pathway. Tumour Biol. 2016;37(5):5885–5895. | ||
Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna-203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int. 2015;15:62. | ||
Gan TQ, Tang RX, He RQ, Dang YW, Xie Y, Chen G. Upregulated MiR-1269 in hepatocellular carcinoma and its clinical significance. Int J Clin Exp Med. 2015;8(1):714–721. | ||
Pan L, Huang S, He R, Rong M, Dang Y, Chen G. Decreased expression and clinical significance of miR-148a in hepatocellular carcinoma tissues. Eur J Med Res. 2014;19:68. | ||
Dang YW, Zeng J, He RQ, Rong MH, Luo DZ, Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15(12):4969–4976. | ||
Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Upsala J Med Sci. 2014;119(1):19–24. | ||
Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PloS One. 2013;8(4):e61054. | ||
Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. | ||
Yin W, Zhao Y, Ji YJ, et al. Serum/plasma microRNAs as biomarkers for HBV-related hepatocellular carcinoma in China. BioMed Res Int. 2015;2015:965185. | ||
Tang G, Shen X, Lv K, Wu Y, Bi J, Shen Q. Different normalization strategies might cause inconsistent variation in circulating microRNAs in patients with hepatocellular carcinoma. Med Sci Monit. 2015;21:617–624. | ||
Wen Y, Han J, Chen J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J cancer. 2015;137(7):1679–1690. | ||
Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55(6):1852–1862. | ||
Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149(5):1226–1239.e1224. | ||
Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21(20):6317–6328. | ||
Wu J, Lv S, An J, Lu C. Pre-miR-149 rs71428439 polymorphism is associated with increased cancer risk and AKT1/cyclinD1 signaling in hepatocellular carcinoma. Int J Clin Exper Med. 2015;8(8):13628–13633. | ||
Shen G, Rong X, Zhao J, et al. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;35(12):2748–2755. | ||
Wang Y, Hu C, Cheng J, et al. MicroRNA-145 suppresses hepatocellular carcinoma by targeting IRS1 and its downstream Akt signaling. Biochem Biophys Res Commun. 2014;446(4):1255–1260. | ||
Wang JG, Zheng XX, Zeng GY, Zhou YJ, Yuan H. Purified vitexin compound 1 induces apoptosis through activation of FOXO3a in hepatocellular carcinoma. Oncol Rep. 2014;31(1):488–496. | ||
Xie C, Xie DY, Lin BL, et al. Interferon-beta gene-modified human bone marrow mesenchymal stem cells attenuate hepatocellular carcinoma through inhibiting AKT/FOXO3a pathway. Br J Cancer. 2013;109(5):1198–1205. | ||
Guo XL, Li D, Hu F, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320(2):171–179. | ||
Liu B, Cao Y, Jiang H, Mao A. Autophagy facilitates the sorafenib resistance of hepatocellular carcinoma cells. West Indian Med J. 2013;62(8):698–700. | ||
Zheng B, Zhu H, Gu D, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459(2):234–239. | ||
Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):835–853. | ||
Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: a comprehensive review. World J Hepatol. 2015;7(26):2648–2663. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




