Back to Journals » OncoTargets and Therapy » Volume 9
Clinical significance of TIPE expression in gastric carcinoma
Authors Hu R, Qiu X, Hong S, Meng L, Hong X, Qiu J, Yang J, Zhuang G, Liu Z
Received 16 November 2015
Accepted for publication 22 May 2016
Published 25 July 2016 Volume 2016:9 Pages 4473—4481
DOI https://doi.org/10.2147/OTT.S100593
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Li
Ruyi Hu,1,* Xingfeng Qiu,1,2,* Shifu Hong,1 Luxi Meng,1 Xinya Hong,3 Jinhua Qiu,1 Jingjing Yang,1 Guohong Zhuang,1,* Zhongchen Liu1,4
1Organ Transplantation Institute, Anti-Cancer Research Center, Medical College, Xiamen University, Fujian, People’s Republic of China; 2Division of Gastroenterology Surgery, Zhongshan Hospital, Gastroenterology Institute of Xiamen University, Gastroenterology Center of Xiamen, Fujian, People’s Republic of China; 3Fujian Medical University, Fujian, People’s Republic of China; 4Department of General Surgery, The Tenth People’s Hospital of Tongji University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Background: TNFAIP8, also known as TIPE, is a suppressor of apoptosis. High expression of both TIPE mRNA and protein has been detected in various cancer cell lines and clinical specimens compared to healthy tissues. Many reports have shown that there is a strong correlation between TIPE overexpression and cancer progression and poor prognosis in human solid cancers.
Methods: To illustrate the functional and clinical significance of TIPE in gastric cancer, we used reverse transcription polymerase chain reaction, quantitative real-time polymerase chain reaction, and immunohistochemistry to measure TIPE expression in clinical gastric specimens. Then, TIPE expression was knocked down by using shRNA and anti-DR5ScFv, to examine different expressions of TIPE in BGC823 cell lines, while cell proliferation and apoptosis were induced.
Results: We found that there was a strong correlation between TIPE expression and TNM stage (P=0.044), tumor depth (P=0.016), lymph node metastasis (P=0.026), and distant metastasis (P=0.045). No significant correlation was found between TIPE expression with the patients’ age (P=0.062) or sex (P=0.459). Anti-DR5ScFv induced TIPE depletion both in vitro and in vivo and resulted in apoptosis and suppression of proliferation.
Conclusion: Our results suggested that TIPE expression was associated with gastric cancer progression, and most importantly, suppressing TIPE expression might be an effective therapeutic strategy.
Keywords: TNFAIP8, gastric cancer, shRNA, anti-DR5ScFv, metastasis, apoptosis
Introduction
Gastric cancer is the most common digestive system tumor. Gastric cancer is lethally aggressive and malignant, so it is necessary to find suitable biomarkers of malignancy to monitor cancer progression. Many mutations and/or abnormal expression of genes are associated with cancer initiation, progression, and metastasis.1
Higher mRNA expression of TNFAIP8, also known as TIPE, GG2-1, SCC-S2, or NDED, has been detected in many malignant cell lines and tissues.2–5 TIPE expression levels have been studied in several kinds of human cancers both in vitro and in vivo, but no study has been conducted in gastric cancer so far. From the current research, studies have shown that TIPE overexpression was found to be correlated with cancer progression and poor prognosis in patients with lung cancer. In contrast, in vitro studies showed the depletion of TIPE mRNA, by using siRNA, in lung cancer cell lines and esophageal squamous cell carcinoma significantly inhibited cell growth and invasion.6,7
The benefit of using small molecules and monoclonal antibodies could be nontoxic to most of the proteins.8 Thus, using siRNA to target mRNA upstream appears to be the next logical strategy. The siRNA can be formed by cutting the hairpin parts of the shRNA by using the Dicer enzyme.9 Instead of directly making siRNA, making the shRNA vectors beforehand showed a more stable, efficient, and long-lasting siRNA.
The aim of this study was to establish the clinical significance and functional features of TIPE expression in gastric cancer. Quantitative real-time polymerase chain reaction (qPCR) and immunohistochemistry were used to evaluate the TIPE expression, and the correlation between TIPE expression and cancer progression was evaluated following the guidelines of REMARK.10 TIPE expression was also knocked down in gastric cancer cell lines (BGC823) by using shRNA, to study the effects of TIPE on cell proliferation and apoptosis.
Materials and methods
Patients and clinical tissue samples
Surgical specimens were obtained from 46 gastric cancer patients (39 males and seven females) who underwent endoscopic biopsy or potentially curative surgery at Zhongshan Hospital, from 2012 to 2014. The patients’ age ranged from 43 to 88 (mean 68) years. Tumor stage and disease grade were classified according to the 6th edition of the tumor-node-metastasis (TNM) classification of the International Union against Cancer. The tumor differentiation evaluation was based on the histological criteria, outlined by the Chinese Society for gastric cancer Disease. None of the patients had received irradiation or chemotherapy before surgery. Patients who had undergone noncurative surgery and/or who had received inadequate follow-up were excluded from the study. Tumor tissues, from which mRNA and protein were isolated, were fresh specimens collected from resection surgery. All samples were obtained after receiving written informed consent from all patients, and the study was approved by the Committee on Medical Ethics of Zhongshan Hospital.
Cell lines and plasmids
The gastric cancer cell lines BGC823 were used. The cell lines were maintained in our laboratory. All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), containing 10% fetal bovine serum and 1% penicillin–streptomycin in a humidified 5% CO2 incubator at 37°C. TIPE shRNA (Top Strand 5′-gatccGCGTTCAGGCACAAAAGAAGTTCAAGAGACTTCTTTTGTGCCTGAACGTTTTTTACGCGTGg-′3, Bottom Strand 3′-gCGCAAGTCCGTGTTTTCTTCAAGTTCTCTGAAGAAAACACGGACTTGCAAAAAATGCGCActtaa-′5) were synthesized in our laboratory. BGC823 were infected with TIPE-shRNA-pSIREN-RetroQ/PVSVG recombinant lentiviral vectors and pSIREN-RetroQ/PVSVG as the negative control lentiviral vectors. Culture medium was replaced 24 hours after infection, and after an additional 48 hours the resistant cell clones were selected with puromycin, the expression of TIPE level was verified by Western blot and reverse transcription PCR (RT-PCR).
Protein extraction and Western blot
All cells were harvested at approximately 80% confluence, and the protein was extracted. The protein concentration was determined using the BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific). The protein (30 μg) samples were run on 12% Tris-Tricine Ready Gel (Bio-Rad Laboratories Inc., Hercules, CA, USA) and then electrotransferred to a Hybond-enhanced chemiluminescence membrane (Amersham). The membranes were incubated overnight at 4°C with rabbit polyclonal antibody against TIPE (1:4,500; Abcam plc, Cambridge, UK) and mouse monoclonal antibody against β-actin (1:5,000; Sigma-Aldrich Co., St Louis, MO, USA). The bands were detected by using an enhanced chemiluminescence detection system, where β-actin served as the control. Scanning densitometry was performed using Adobe Photoshop (Apple, Inc, Cupertino, CA, USA).
RNA extraction and RT-PCR
Total RNA was extracted from the tissue using the TRIzon total RNA extraction reagent (CWBIO, Beijing, People’s Republic of China). RNA quality and concentration were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Reverse transcription and PCR were performed using Takara PrimeScript™ RT-PCR Kit (Takara-Dalian, Dalian, People’s Republic of China) and 2× EasyTaq® PCR super mix (TransGen, Beijing, People’s Republic of China), template cDNA was synthesized from 2.0 μg of total RNA. RT-PCR was performed using the 96-well Biometra PCR System in a total volume of 20 μL at 94°C for 3 minutes followed by 30 cycles of 94°C for 30 seconds, 56°C for 30 seconds, 72°C for 1 minute, and 72°C for 5 minutes. The sequences of the primer pairs were as follows: TIPE forward, 5′-CCCAGGGAAGTGGCTACAGA-′3, TIPE reverse, 5′-GCCTCCTTCTTGTTCTGGGT-′3 and β-actin forward, 5′-AGCCATGTACGTAGCCATCC-′3, β-actin reverse, 5′-ACCCTCATAGATGGGCACAG-′3. RT-PCR imaging results, application of gel analysis, and the gray value against the corresponding β-actin were used as the reference.
qRT-PCR
cDNA synthesis and qPCR were performed using ReverTra Ace® qPCR RT Kit and SYBR® Green Realtime PCR Master Mix Plus (TOYOBO, Osaka-shi, Japan), respectively. The StepOne Real-Time PCR System (ABI, Cambridge, UK) was used to perform the reaction, where β-actin was used to normalize data. The data of the qPCR were calculated using the 2−ΔΔC(t) method.
Expression and purification of human anti-DR5ScFv (ZF1)
The obtained bacterial recombination was selected and cultured at 37°C in 3 mL Luria–Bertani broth (LB medium) for 8–12 hours, later it was added to 300 mL of fresh LB medium at 37°C/250 rpm. After 7 hours, when the cell density reached between 0.6–0.8 (OD590) isopropyl-β-D-thiogalactopyranoside was added to LB medium to induce expression. Escherichia coli Rosetta-gami™ (EMD Millipore, Billerica, MA, USA) cells from shaker flasks were disintegrated with ultrasonication (300 W, 30 seconds per/30-second intervals for 20 minutes). The suspension was precipitated by centrifugation (12,000 rpm, 20 minutes). The target protein was mainly present in inclusion bodies. The inclusion bodies were washed at 4°C for 7–8 hours in a washing fluid containing 2 M urea. After washing, the inclusion bodies were dissolved in a solution containing 8 M urea. The solution was centrifuged, and the supernatant was transferred to a Ni-nitrilotriacetic acid affinity column (GE Healthcare Life Sciences, Piscataway, NJ, USA) to purify the protein. After loading and eluting, the targeted fractions were pooled and identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Immunohistochemistry
Tumor tissues were fixed and stained following the methods used in Yang et al’s work.11 And the concentration of affinity-purified rabbit TIPE antibody (Ab) was 1.67 g/mL.
Apoptosis assay by flow cytometry
To assess apoptosis, cells were plated at a concentration of 5×105 cell/L in the six-well plate, incubated for 24 hours, then treated with ZF1 (0.1 mg/mL) for 8 hours. The control groups were incubated in Dulbecco’s Modified Eagle’s Medium only. After incubation, the cells were harvested with 0.25% trypsin (without ethylenediaminetetraacetic acid), then washed with phosphate-buffered saline and further suspended in a binding buffer (1×). An aliquot of 100 μL cell suspension was incubated with 5 μL of Annexin V-APC and 5 μL of propidium iodide (PI) for 15 minutes in the dark, then 1 mL binding buffer (1×) was added to each sample. The cells were analyzed directly by a flow cytometer. The percentage of cells was calculated by FlowJo Software.
TUNEL assay
To detect apoptosis in in vivo treatments, we used the TUNEL method. The apoptotic cells are defined by the uptake of PI (red) and FITC (green) (indicating the damage to the cell membrane) and the presence of clear nuclear condensation and/or fragmentation with DNA ends of dUTPs. The paraffin-embedded sections were deparaffinized in xylene and rehydrated in a graded series of ethanol baths. The sections were treated with 20 g/mL of proteinase K in distilled water for 10 minutes at room temperature. The tumor and liver tissues were fixed in 1% paraformaldehyde for 10 minutes. To block endogenous peroxidase, the slides were incubated in methanol containing 0.3% hydrogen peroxide for 20 minutes. The remaining procedures were performed according to the instructions provided by the Roche company. For quantification of apoptosis, five microscopic fields were randomly selected at high power magnification (200×), and the average counts of TUNEL-positive cells were calculated.
TIPE knock-down in a nude mice tumor model
We divided the nude mice into four groups, each group consisted of six animals. All four groups received the injection of BGC823 cells at a dose of 1×107 cells per mouse; however, two groups received the cells which were treated with shRNA and the other two groups received nontreated cells, as the control. When the tumor nodules started to grow it was classified as day 0. On day 3, each one of the shRNA-treated and the control group received the ZF1 injections, 80 mg/kg every day. The other two groups received saline injections. Tumor length (L), width (W), and diameter were measured every 3 days; tumor volume (mm3) was calculated using the formula W2 × (L/2). The mice were sacrificed on day 14, and the neutral formalin-fixed tumor samples were stained with hematoxylin–eosin (H&E), and TUNEL assay was also conducted as mentioned previously. All experimental procedures were conducted in conformity with institutional guidelines for the care and use of laboratory animals, and protocols were approved by the Animal Studies Committee of Xiamen University, People’s Republic of China.
H&E staining analysis
To distinguish antitumor effects of ZF1 in vivo, H&E staining was used. Tumor and liver tissues were harvested (n=6). The H&E stained tissues were used to identify necrotic cells, tumor cells or apoptotic cells. H&E stained sections were viewed under an Olympus BHT microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Data were presented as mean ± standard deviation. Statistical analysis was performed for multiple comparisons using analysis of variance and Student’s t-test. A value of P<0.05 was considered to be statistically significant.
Results
Expression of TIPE protein and mRNA in tumor cell lines and clinical gastric tissue specimens
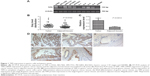
We examined TIPE protein and mRNA expression in human gastric cancer cell lines (MGC803, BGC823), human leukemia cell line (K562), human hepatoma cancer cell line (SMC7721), human colon cancer cell line (SW480), and human gastric cancer clinical specimens with different tumor grades using the RT-PCR method. The expression level of TIPE protein and mRNA was all statistically significant in all these tested samples (Figure 1A). Based on the earlier results, we used the BGC823 cell line to perform TIPE suppression analysis. We found that the TIPE mRNA was not only expressed in gastric cancer tissues but also in the tested adjacent gastric tissues.
The expression of TIPE was investigated by using RT-PCR in 46 gastric cancer specimens. The results of TIPE mRNA expression in a gastric tumor and its adjacent tissues are shown in Figure 1B. Our results showed that TIPE mRNA expression in gastric cancer tissue was statistically higher (P<0.05) than in adjacent tissues; to further confirm TIPE mRNA expression level, qPCR was used (Figure 1C). The mean expression value of TIPE mRNA level in cancer tissues (1.417±0.342) was significantly higher than the value (0.404±0.091) in normal gastric cancer tissues (P<0.05). According to our results obtained from RT-PCR and qPCR, TIPE mRNA expression levels showed positive correlations to tumor invasion in gastric cancer.
The correlations between TIPE mRNA expression level and the clinicopathological characteristics of gastric cancer patients (age, sex, differentiation, TNM stage, lymph node metastasis, distant metastasis, lymphatic invasion, and venous invasion) are shown in Table 1. There were significant correlations between TIPE expression and TNM stage (P=0.044), tumor depth (P=0.016), lymph node metastasis (P=0.026), and distant metastasis (P=0.045). No significant correlations were found between TIPE expression and age (P=0.459) and sex (P=0.062). In univariate analysis, high TIPE expression was found to be a significant prognostic factor for poor survival, in addition to tumor depth and differentiation, the presence of lymph node metastasis and distant metastasis.
  | Table 1 The expression of TIPE in gastric carcinoma and their clinicopathological relationship |
Overexpressed TIPE was detected in 46 gastric cancer samples, whereas negative or weak TIPE immunoreactivity was observed in adjacent tissues. TIPE was clearly localized in the cytoplasmic compartment of the tumor cells (Figure 1D). Finally, we found that the expression of TIPE in poorly differentiated gastric adenocarcinoma was higher compared with that in well-differentiated ones, a similar difference was found between adenocarcinoma stages II and III.
Depletion of TIPE expression inhibits tumor cell growth in gastric cancer cell lines
Higher expression levels of TIPE contribute to the proliferation of cancer cells. We therefore hypothesized that decreased expression of TIPE might lead to cell death.
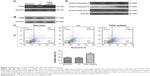
Firstly, to test our hypothesis, we transfected BGC823 cells with a TIPE knock-down plasmid and maintained BGC823 stable transfectants (TIPE shRNA and vector) in puromycin-containing medium. Then, we confirmed the expression of endogenous TIPE (21 kDa) by using Western blotting and RT-PCR (132 bp). Western blot and RT-PCR analysis confirmed that TIPE shRNA significantly reduced the expression of TIPE (Figure 2A and B).
Apoptosis was examined by using flow cytometry after staining the cells with annexin-V and PI. As a result, BGC823 cells were treated with ZF1, 19.40% of cells underwent apoptosis, 17.98% were found in negative control cells; and 31.68% in the ZF1 treated shRNA-BGC823 cells (Figure 2C). Results showed ZF1 was able to induce more apoptosis in the TIPE treated shRNA group.
Analysis of the apoptosis pathway associated protein expression level revealed that the levels of activated caspase-8/3/9 were different in each group (Figure 2D). In comparison, the activated caspase-8/3/9 levels were similar in both negative control groups, which were all treated with ZF1, whereas activated caspase-8/3/9 levels were elevated in the TIPE shRNA treated BGC823 cells. These results, gathered from TIPE shRNA treated BGC823 cells, confer the susceptibility to ZF1 induced apoptosis in the absence of TIPE. These results indicated that TIPE might be involved with negative regulation of cell death. This inhibition could possibly be associated with caspase activity.
Knock-down TIPE impairs gastric cancer formation of BGC823 cells in vivo and enhanced anti-tumor effect of anti-DR5ScFv in tumor-bearing athymic mice
To verify the correlation of TIPE expression with experimental tumor growth and drug resistance, a xenograft tumor model was established by injecting nude mice with BGC823 cells and TIPE shRNA treated BGC823 cells. We examined the effects of inhibition of endogenous TIPE expression on the xenograft tumor growth of BGC823 cells and the susceptibility to ZF1.
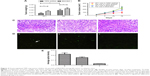
Measurement of tumor size showed that at 14 days, tumors induced by TIPE shRNA treated BGC823 cells (0.019±0.004 g, 232.586±31.632 mm3) were significantly smaller than tumors from the control group (0.026±0.003 g, 252.825±34.883 mm3, P<0.05). Tumors of TIPE shRNA transfected BGC823 cells treated with ZF1 (0.008±0.002 g, 94.987±24.287 mm3) were smaller than tumors treated with saline (0.016±0.003 g, 178.387±8.083 mm3, P<0.05) (Figure 3A and B).
There were no statistical differences in weight of mice between the ZF1 and saline groups (P>0.05). Furthermore, all the mice were observed to be in a healthy condition and remained active throughout the study, suggesting that ZF1 administration had no obvious physiological or psychological side effects. H&E staining revealed that TIPE shRNA promotes more tumor cell death in vivo than tumor tissue from nude mice injected with ZF1, as well as disappearance of intracellular structures and cell sputtering (Figure 3C).
In contrast, tumor tissue from control nude mice injected with saline showed large and irregular cell shapes as expected. Results obtained from TUNEL stain showed apoptosis positive cells were more abundant in tumor tissue from TIPE shRNA mice than in tissue from the two control groups (Figure 3D and 3E).
Discussion
TIPE was originally identified in a primary human head and neck squamous cell carcinoma cell line; with its metastatic cell line established from cancer recurrence after radiation therapy.12 Overexpression of TIPE in cancer cells enhances cell survival and proliferation in various cells.2,13,14 High expression of TIPE in cancer cells is correlated with enhanced cell survival and inhibition of apoptosis, including inhibition of caspase-8 and caspase-3.2,15
This is the first report on the role of TIPE in gastric cancer. TIPE overexpression was significantly associated with high histologic grade, large residual tumor size, recurrence, and poor response to chemotherapy.16 Our data demonstrated that expression of TIPE protein was much higher in several human cancer cells; MGC803, BGC823, K562, SMC7721, and SW480. These results were in line with previous published data; TIPE was discovered to have higher expression levels in various human cancer cell lines, such as K562, MOLT4, A549, and SKOV-3, but lower levels in SW480.15 Moreover, TIPE expression was found to be higher in gastric carcinomas compared to the normal adjacent tissues examined. In this study, results showed significant correlations between TIPE expression and TNM stage (P=0.044), tumor depth (P=0.016), lymph node metastasis (P=0.026), and distant metastasis (P=0.045). No significant correlations were found between TIPE expression and age (P=0.062) or sex (P=0.459). Its expression pattern is similar to human breast carcinomas, non-small cell lung cancer, and renal cell carcinomas.4,6,17
In the absence of TIPE expression in BGC823 cells, via shRNA knock-down, there was increased cell death in BGC823 cells. Our findings of decreased TIPE expression and increased cell death are consistent with the role of TIPE in cell survival and metastasis.13,15
Anti-DR5mAb could induce cell apoptosis by binding to DR5. He et al’s, group showed that apoptosis plays a key role in antitumor activities in colon cancer cells and xenografts through the DR5, FADD, and caspase-8 axis.18 Overexpression of DR5 and DR4 receptors can induce tumor cell apoptosis and reverse the drug resistance of tumors to TRAIL.19 Activated DR5 recruits the adaptor protein FADD, then cleaves procaspase-8 and initiates caspase-8 activation, which activates the downstream effector caspase-3 that leads to apoptosis.20,21
Our laboratory successfully prepared and purified a high purity and stable Anti-DR5ScFv, which could specifically bind to eDR5 and induce cell apoptosis.11 TIPE could suppress TNF-α-mediated apoptosis by inhibiting caspase-3 and caspase-8 activity.2,17 This inhibition might lead to its resistance to chemotherapy and antibody treatments, thereby halting tumor progression.
Conclusion
In this study, we demonstrated that after the administration of Anti-DR5ScFv, apoptosis was induced to a greater degree in the TIPE shrank group than in the control group, both in vitro and in vivo. In addition, the activity of caspase-8, caspase-3, and caspase-9 was successfully induced, and more apoptotic cells were found in the tumor in the absence of TIPE. Therefore, TIPE might play a critical role in interfering with anticancer drug resistance. In the absence of TIPE expression in BGC823, induced cell apoptosis was significantly elevated, showing a direct link between TIPE expression and tumor cell growth inhibition. These findings indicated that TIPE would be a useful tumor therapy target in the close future.
Acknowledgments
This work was supported by grants obtained from the National Natural Science Foundation of China (No 81272720), Fujian province (No 3402Z20100002), Medical innovation project Fujian province (2014-CXB-43), Public Projects of Fujian Province 2016R1034-3, and key projects for technology plan of Xiamen city.
The authors thank Chunyan Shi, Wenming Liu, Lihong Chen, Cailing Tong, and Zhongquan Qi for their help and advice during this research.
Disclosure
The authors report no conflicts of interest in this work.
References
Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13(6):464–477. | ||
You Z, Ouyang H, Lopatin D, Polver PJ, Wang CY. Nuclear factor-kappa B-inducible death effector domain-containing protein suppresses tumor necrosis factor-mediated apoptosis by inhibiting caspase-8 activity. J Biol Chem. 2001;276(28):26398–26404. | ||
Zhang C, Chakravarty D, Sakabe I, et al. Role of SCC-S2 in experimental metastasis and modulation of VEGFR-2, MMP-1, and MMP-9 expression. Mol Ther. 2006;13(5):947–955. | ||
Kumar D, Gokhale P, Broustas C, Chakravarty D, Ahmad I, Kasid U. Expression of SCC-S2, an antiapoptotic molecule, correlates with enhanced proliferation and tumorigenicity of MDA-MB 435 cells. Oncogene. 2004;23(2):612–616. | ||
Zhang S, Zhang Y, Wei X, et al. Expression and regulation of a novel identified TNFAIP8 family is associated with diabetic nephropathy. Biochim Biophys Acta. 2010;1802(11):1078–1086. | ||
Dong QZ, Zhao Y, Liu Y, et al. Overexpression of SCC-S2 correlates with lymph node metastasis and poor prognosis in patients with non-small-cell lung cancer. Cancer Sci. 2010;101(6):1562–1569. | ||
Hadisaputri YE, Miyazaki T, Suzuki S, et al. TNFAIP8 overexpression: clinical relevance to esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19 Suppl 3:S589–S596. | ||
de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6(6):443–453. | ||
Vermeulen A, Behlen L, Reynolds A, et al. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11(5):674–682. | ||
McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387–391. | ||
Yang J, Huang X, Luo F, et al. Preparation and functional studies of hydroxyethyl chitosan nanoparticles loaded with anti-human death receptor 5 single-chain antibody. Onco Targets Ther. 2014;7:779–787. | ||
Patel S, Wang FH, Whiteside TL, Kasid U. Identification of seven differentially displayed transcripts in human primary and matched metastatic head and neck squamous cell carcinoma cell lines: implications in metastasis and/or radiation response. Oral Oncol. 1997;33(3):197–203. | ||
Zhang HG, Hyde K, Page GP, et al. Novel tumor necrosis factor alpha-regulated genes in rheumatoid arthritis. Arthritis Rheum. 2004;50(2):420–431. | ||
Horrevoets AJ, Fontijn RD, van Zonneveld AJ, de Vries CJ, ten Cate JW, Pannekoek H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood. 1999;93(10):3418–3431. | ||
Kumar D, Whiteside TL, Kasid U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin-1beta-converting enzyme-inhibitory protein. J Biol Chem. 2000;275(4):2973–2978. | ||
Liu T, Gao H, Chen X, et al. TNFAIP8 as a predictor of metastasis and a novel prognostic biomarker in patients with epithelial ovarian cancer. Br J Cancer. 2013;109(6):1685–1692. | ||
Laliberte B, Wilson AM, Nafisi H, et al. TNFAIP8: a new effector for Galpha(i) coupling to reduce cell death and induce cell transformation. J Cell Physiol. 2010;225(3):865–874. | ||
He K, Zheng X, Li M, Zhang L, Yu J. mTOR inhibitors induce apoptosis in colon cancer cells via CHOP-dependent DR5 induction on 4E-BP1 dephosphorylation. Oncogene. 2016;35(2):148–157. | ||
Liu X, Yue P, Khuri FR, Sun SY. p53 upregulates death receptor 4 expression through an intronic p53 binding site. Cancer Res. 2004;64(15):5078–5083. | ||
Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12(6):611–620. | ||
Suliman A, Lam A, Datta R, Srivastava RK. Intracellular mechanisms of TRAIL: apoptosis through mitochondrial-dependent and -independent pathways. Oncogene. 2001;20(17):2122–2133. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.