Back to Journals » International Journal of Nephrology and Renovascular Disease » Volume 12
Clinical Significance Of Linc00342 Expression In The Peripheral Blood Lymphocytes Of Patients With Chronic Kidney Disease
Authors Liu C, Xu Y, Wu X , Zou Q
Received 24 March 2019
Accepted for publication 29 October 2019
Published 12 December 2019 Volume 2019:12 Pages 251—256
DOI https://doi.org/10.2147/IJNRD.S209832
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pravin Singhal
Cheng Liu,1 Yaqian Xu,1 Xueping Wu,2 Qi Zou1
1Department of Critical Care Medicine, The First Affiliated Hospital of Bengbu Medical College, Bengbu, Anhui 233004, People’s Republic of China; 2Department of Nephrology, The First Affiliated Hospital of Bengbu Medical College, Bengbu, Anhui 233004, People’s Republic of China
Correspondence: Xueping Wu
Department of Nephrology, The First Affiliated Hospital Of Bengbu Medical College, Bengbu, Anhui 233004, People’s Republic of China
Email [email protected]
Qi Zou
Department of Critical Care Medicine, The First Affiliated Hospital of Bengbu Medical College, Bengbu, Anhui 233004, People’s Republic of China
Email [email protected]
Objective: To investigate the expression of Linc00342 in peripheral blood lymphocytes in patients with chronic kidney disease (CKD) and healthy people and to identify Linc00342 as a biomarker of chronic kidney disease.
Methods: Peripheral blood samples were collected from 30 patients with chronic kidney disease and 10 healthy volunteers at the First Affiliated Hospital of Bengbu Medical College, China. According to CKD classification, the patients were divided into three CKD groups (the CKD1/2 group, CKD3/4 group and CKD5 group) and a healthy volunteer group (the H group). The relative expression of Linc00342 in lymphocytes was detected by RT-PCR, while the IL-6 and IL-10 levels in the serum were detected by ELISA. In addition, the general data of the patients and healthy volunteers were recorded. Finally, SPSS software was used for statistical analysis.
Results: The expression level of Linc00342 in the peripheral blood lymphocytes of the four groups increased significantly as the CKD grade increased, and there were statistically significant differences (p < 0.01). There was a positive linear correlation between the expression of Linc00342 in peripheral blood lymphocytes and the eGFR (p < 0.05), which was expressed by the linear model equation: Y = 2.532 + 0.012X. Among the inflammatory factors for the early diagnosis of CKD, the area under the ROC curve for the expression of Linc00342 in peripheral blood lymphocyte was 0.953, the standard error was 0.034 and the 95% confidence interval was 0.000–1.000.
Conclusion: The expression level of Linc00342 in peripheral blood lymphocytes can reflect the severity of CKD, and Linc00342 is possibly used as a molecular marker of CKD.
Keywords: Linc00342, chronic kidney disease, lymphocyte
Introduction
The etiology of chronic kidney disease (CKD) is complex, including primary glomerular disease, secondary glomerular disease, hereditary glomerular disease and chronic tubulointerstitial damage caused by various factors, and chronic nephritis, lupus nephritis, diabetic nephropathy and polycystic kidney disease are common types of CKD.1 Currently, the analysis of blood from CKD patients mainly focuses on metabolomics, and more than 90 molecular compounds have been found to be related to the severity of CKD.2,3 However, the molecular mechanisms of specific pathogenic processes have not yet been elucidated. Some studies have identified some microRNAs, lncRNAs and genes related to CKD as well as their biological functions and pathways through expression microarray experiments using blood samples from CKD patients, and these studies have laid a foundation for the study of the molecular mechanisms of CKD.
In our previous work,4 we searched the gene expression database (GEO) of the National Center for Biotechnology Information (NCBI) of the United States and collected a set of genome-wide expression profiles from the peripheral blood of 75 CKD patients and 20 normal subjects (data number: GSE37171). The chip experimental platform that we used was the Affymetrix Human Genome U133 Plus 2.0 Array (platform number: GPL570). The robust multiarray average (RMA) algorithm was used to standardize the expression spectrum of the original data (the analysis software was Affymetrix expression console software). Then, CKD-related differentially expressed genes (| log 2 (fold change) | >1; FDR < 0.05) were screened by the Significance Analysis of Microarrays (SAM) algorithm (the analysis platform was the R language platform).5 The results showed that Linc00342 was differentially expressed between the peripheral blood of CKD patients and that of healthy people (Table 1). All coexpressed genes were mapped using Cytoscape software to form a gene coexpression network. Then, DAVID software was used to enrich and analyze the functions of the genes (adjusted P < 0.05). Genes in the Linc00342-related coexpression network were mainly enriched in biological functions and pathways related to apoptosis, the inflammatory response and protein hydrolysis (Figure 1). The purpose of this study was to further understand the clinical significance of Linc00342 expression in CKD by verifying the differential expression of Linc00342 between the peripheral blood of CKD patients and that of healthy people.
 |
Table 1 Linc00342 With Differential Expression In Peripheral Blood Of Chronic Kidney Disease |
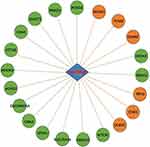 |
Figure 1 Linc00342 co-expression network. Red and green nodes represent upregulated and downregulated genes, respectively. |
Materials And Methods
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College, China (approval number: BYYFY-2017KY07). Informed consent was obtained from all conscious patients and healthy volunteers enrolled in the study. We first verbally informed the detailed informed consent, including the experimental significance, purpose, methods and possible hazards, and then signed the written informed consent. If the patients were conscious, the written informed consent was signed by the patients and their first guardians. If the patients were unconscious, the written informed consent was signed by their first guardians.
The subjects were 30 patients with CKD diagnosed for the first time in the Department of Nephrology of the First Affiliated Hospital of Bengbu Medical College, China between March 2018 and December 2018 and 10 healthy volunteers (group H) who were selected from the physical examination center. According to the staging of CKD (Improving Global Outcomes [KIDGO] clinical practice guidelines: stage 1, eGFR>90; stage 2, 60<eGFR<90; stage 3, 30<eGFR<60; stage 4, 15<eGFR<30; stage 5, eGFR<15; [unit:mL/min/1.73m2]), the patients with CKD were divided into three groups: CKD1/2 group, CKD3/4 group and CKD5 group.6 The exclusion criteria were as follows: 1) age <18 years or >70 years; 2) presence of tumors; 3) presence of infection; 4) dialysis use; 5) pregnant and postpartum women; 6) presence of serious complications and 7) hormone therapy or immunosuppressive agent use.
Five milliliters of peripheral blood from the patients with CKD and healthy volunteers was used to detect serum creatinine (Cr). According to age and sex, the eGFR was estimated by the CKD-EPI formula,7 i.e., eGFR=a×(Cr/b)c×0.993 age, [a=144 (female) or 141 (male), b=0.7 (female) or 0.9 (male), c=−0.329 (female, Cr<0.7 mg/dL) or −1.209 (female, Cr>0.7 mg/dL) or −0.413 (male, Cr<0.7 mg/dL) or −1.209 (male, Cr>0.7 mg/dL)].
Eight milliliters of peripheral blood from the patients with CKD and healthy volunteers was used to isolate lymphocytes and the serum, which was kept in a −80°C freezer. Then, ELISAs were used to detect the expression of IL-6 and IL-10. RT-PCR was used to detect the relative expression of Linc00342.
Experimental Reagents
Reagents for the detection of IL-6 and IL-10 in the serum by ELISA were purchased from R&D Company (USA). The following reagents were purchased: a lymphocyte separation solution (TBD LTS 1077, Tianjin Bayang Biological Products Technology Co., Ltd.), RNeasy Plant Mini Kit (TIANGEN DP501, Beijing Tiangen Biochemical Technology Co., Ltd.), the RevertAid First Strand cDNA Synthesis Kit K1622 (Thermo Fisher Scientific) and RR820A takaraTB Green™ Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio). Primers for Linc00342 and GAPDH, which were used as the internal reference gene, were synthesized by Bioengineering (Shanghai) Co., Ltd. The Linc00342 primer sequences were Linc00342-F CCCAAAGCAGTCCTTCACTACA and Linc00342-R CTGCAGTTCACTCTGCTGCTT.
IL-6 and IL-10 were detected by ELISAs, which were conducted strictly according to the instructions of the reagent manufacturer.
Peripheral Blood Lymphocyte Extraction
A lymphocyte separation solution (TBD LTS 1077) was added into a centrifuge tube. Venous blood with the anticoagulant heparin was mixed with an equal amount of saline. A dropper was used to slowly superimpose the blood on the lymphocyte separating solution, and a clear interface was maintained. The tube was horizontally centrifuged at 2000 rpm for 20 mins. After centrifugation, the tube was divided into three layers, including an upper layer containing the plasma and saline, and a lower layer mainly containing erythrocytes and granulocytes. The middle layer contains the lymphocyte separation fluid, and there is a narrow, white layer dominated by mononuclear cells at the upper and middle interfaces. Capillary tubes were inserted into the white layer to absorb the mononuclear cells. Then, the cells were washed twice by inserting another short tube, adding an amount of saline that was more than 5 times the tube volume, and centrifuging at 1500 rpm for 10 mins.
Total RNA Extraction From Lymphocytes And RT-PCR
Total RNA was extracted from lymphocytes by TIANGENDP501. After the RNA was extracted, the concentration of the RNA was determined by using a NanoDrop 1000 spectrophotometer, and the expression of Linc00342 was determined by using a 7900HT fluorescence quantitative PCR system. Through threshold analysis and comparison, the data were analyzed and processed by the cyclic number (ct) method.
Statistical Methods
SPSS 18.0 was used for statistical analysis. Normal distribution measurements were expressed in terms of x± s. Variance analysis was used for comparisons between groups. The χ2 test was used to compare the counting data between groups. The Pearson method was used for correlation analysis, and ROC curves were used to determine the sensitivity and specificity of IL-6, IL-10 and Linc00342 for the diagnosis of CKD. Differences were considered statistically significant at P < 0.05 and P < 0.01.
Results
There were no significant differences in age, sex or primary disease among the four groups (p > 0.05). There were significant differences in the creatinine level, eGFR, electrolyte concentration (K+, Ca2+, P and HCO3−), IL-6 level and IL-10 level among the four groups (p < 0.01), as shown in Table 2.
 |
Table 2 Baseline Characteristics In Health Volunteers And CKD Patients |
The expression level of Linc00342 in the peripheral blood lymphocytes of the four groups increased significantly as the CKD grade increased (p < 0.01), as shown in Figure 2.
There was a positive linear correlation between Linc00342 expression and the eGFR in peripheral blood lymphocytes (p < 0.05). Through linear regression analysis, the linear model equation between Linc00342 and eGFR was established to be Y = 2.532 + 0.012X, as shown in Figure 3.
 |
Figure 3 Establishment of linear relation and linear regression model between expression levels of Linc00342 in peripheral blood lymphocyte and eGFR. |
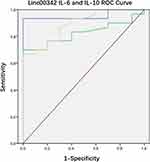
The area under the ROC curve of Linc00342 expression in peripheral blood lymphocytes was 0.953, the standard error was 0.034 and the 95% confidence interval was 0.000–1.000. The area under the ROC curve of the IL-6 level in the serum was 0.823, the standard error was 0.064 and the 95% confidence interval was 0.698–0.949. The area under the ROC curve of the IL-10 level in the serum was 0.890, the standard error was 0.053 and the 95% confidence interval was 0.787–0.993, as shown in Figure 4.
Discussion
A gene is the basic structure and function unit of genetic information, and genes control all aspects and processes of an organism’s life activities and interact with the external environment to regulate various physiological and biochemical processes in life. Therefore, genes are also the basic factors that ensure that biological health is not affected by disease. Most of the biological signaling pathways related to various life activities discovered by humans are composed of various gene networks. Of course, in the courses of various diseases, the most direct participants are also genes. The role of genes is to be transcribed into various RNA molecules, such as mRNA, microRNA, snRNA and lncRNA. At present, many studies have been carried out to examine genes related to chronic kidney disease, among which transforming growth factor-β 1 (TGF-β1) is the main regulator of the ECM and one of the main mediators of renal fibrosis.8 Zhou et al found that FNA1 gene mutation occurs in patients with a specific type of nephropathy, megakaryocytic interstitial nephropathy (KIN), and confirmed the causal relationship between FAN1 mutation and this disease.9 Lebherz et al found that keratin 18 (K-18) and apoptotic protein CK18 (ccK-18) are highly correlated with chronic kidney disease. These two genes are involved in cell apoptosis and necrosis in the pathogenesis of chronic kidney disease.10 Tan et al found that genes in the Wnt/beta-catenin signaling pathway can promote the course of chronic kidney disease by regulating downstream fibrosis-related factors.11
LncRNAs play important roles in life activities and various intracellular signaling pathways and are a hotspot in genetic research. LncRNA is a kind of RNA molecule whose transcript length exceeds 200 nt, and lncRNAs cannot be translated into proteins. LncRNA expression is tissue-specific and varies in different stages of life activities. It is believed that lncRNAs also play key regulatory roles in cells and are related to the basic functions of life activities and diseases.12 Some scholars found that the lncRNA MALAT1 increases the expression of SAA3 in the kidneys of diabetic mice. Silencing MALAT1 could normalize the mRNA levels of SAA3, IL-6 and TNF.13 Yi et al found that linc-Gm4419 is associated with NF-kappa B in the kidney tissue of db/db mice with diabetic nephropathy. If the linc-Gm4419 gene was knocked out, the expression of proinflammatory cytokines and renal fibrosis markers in mesangial cells in a high-glucose environment would be significantly reduced, and cell proliferation would also be reduced.14
In our previous work, we found that Linc00342 is differentially expressed between the peripheral blood of CKD patients and that of healthy people, and Linc00342 is related to inflammation/apoptosis-related functions and pathways.4 Studies have shown that Linc00342 and P53 gene deletions are associated with adverse prognostic markers in small cell lung cancer.15 This study also verified the differential expression of Linc00342 between the peripheral blood lymphocytes of healthy volunteers and those of patients with chronic kidney disease (p < 0.01) and showed that Linc00342 expression gradually increased with increasing CKD grade, consistent with the findings for creatinine, the eGFR and IL-6. Further analysis showed that the expression of Linc00342 in peripheral blood lymphocytes was positively correlated with the eGFR (p=0.045), suggesting that the level of Linc00342 could reflect the condition of the eGFR and was related to disease severity. IL-6 and IL-10 are the key mediators of inflammation in chronic kidney disease. Linc00342, IL-6 and IL-10 are sensitive and specific for the diagnosis of chronic kidney disease. The analysis of the ROC curves for Linc00342, IL-6 and IL-10 show that the Linc00342 curve has higher sensitivity and a larger area under the curve (0.953 vs 0.823 vs 0.890, respectively), suggesting that the increase in the Linc00342 level in peripheral blood lymphocytes is more useful for the diagnosis of CKD.
Previous studies had shown that the CKD patients had lymphatic fineness Cytopenia, and decreased lymphocyte counts occurred long before the end stage of renal disease.16 The reason for T cell decrease may be mainly related to T lymphocyte apoptosis.17 In the previous study of this paper, we found that the function of Linc00342 is related to apoptosis by the enrichment analysis of the genes in Linc00342 related co-expression networks. These results suggest that the increased expression of Linc00342-related genes possibly be related to the increase of lymphocyte apoptosis. The next step is to study the concrete relationship between them.
In summary, this study suggests that studying the mechanism and characteristics of chronic kidney disease at the lncRNA level may lead to a new perspective and new ideas for understanding chronic kidney disease; that is, Linc00342 may become a preventive marker and potential therapeutic target in chronic kidney disease and provide a new approach for the prevention and treatment of chronic kidney disease.
Funding
The project is supported by the project fund of Bengbu Medical College (BYKY1741ZD).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Meyer TW. Hostetter TH: approaches to uremia. J Am Soc Nephrol. 2014;25(10):2151–2158. doi:10.1681/ASN.2013121264
2. Kestenbaum B, Belozeroff V. Mineral metabolism disturbances in patients with chronic kidney disease. Eur J Clin Invest. 2007;37(8):607–622. doi:10.1111/eci.2007.37.issue-8
3. Meyer TW. Hostetter TH: uremia. N Engl J Med. 2007;357(13):1316–1325. doi:10.1056/NEJMra071313
4. Cheng L. Exploring Molecular Mechanism of Uremia by Analysis the Transcriptome Expression Data of Peripheral Blood in Uremia Patients. Anhui Medical University; 2017:1–75.
5. Cheng L, Yonggui W. Co-expression analysis of blood cell genome expression to preliminary investigation of regulatory mechanisms in uremia. Med Sci Monit. 2017;23:38–45. doi:10.12659/MSM.899385
6. Webster AC, Nagler EV, Morton RL. Chronic kidney disease . Lancet. 2012;379(9811):165–180. doi:10.1016/S0140-6736(11)60178-5
7. National Kidney Foundation. KDOQI clinical practice guideline for diabetes and CKD:2012 update. Am J Kidney Dis. 2013;61(6):1049.
8. Meng XM, Chung AC, Lan HY. Role of the TGF-β/BMP-7/Smad pathways in renal diseases. Clin Sci. 2013;124:243–254. doi:10.1042/CS20120252
9. Zhou W, Otto EA, Cluckey A, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44(8):910. doi:10.1038/ng.2347
10. Lebherz-Eichinger D, Krenn CG, Roth GA. Keratin 18 and heat-shock protein in chronic kidney disease. Adv Clin Chem. 2013;62:123–149.
11. Tan RJ, Zhou D, Zhou L. Wnt/β-catenin signaling and kidney fibrosis. Kidney Int Suppl. 2014;4(1):84–90. doi:10.1038/kisup.2014.16
12. Sun M, Kraus WL. From discovery to function:the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64.
13. Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. Cel. Mol Med. 2015;19:1418–1425. doi:10.1111/jcmm.12576
14. Yi H, Peng R, Zhang LY, et al. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8(2):e2583. doi:10.1038/cddis.2016.451
15. Tang H, Zhao L, Li M, Li T, Hao Y. Investigation of LINC00342 as a poor prognostic biomarker for human patients with non-small cell lung cancer. Cell Biochem. 2018;15:10–19.
16. Xiang FF, Zhu JM, Cao XS, et al. Lymphocyte depletion and subset alteration correlate to renal function in chronic kidney disease patients. Ren Fail. 2016;38(1):7–14. doi:10.3109/0886022X.2015.1106871
17. Masumoto Y, Shinzato T, Amano I, et al. Relationship between susceptibility to apoptosis and Fas expression in peripheral blood T cells from uremic patients:a possible mechanism for lymphopenia in chronic renal failure. Biochem Biophys Res Commun. 1995;215(1):98. doi:10.1006/bbrc.1995.2438
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.