Back to Journals » OncoTargets and Therapy » Volume 10
Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: a comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases
Authors Xie ZC, Dang YW, Wei DM, Chen P, Tang RX, Huang Q, Liu JH, Luo DZ
Received 13 March 2017
Accepted for publication 30 May 2017
Published 10 August 2017 Volume 2017:10 Pages 3991—4005
DOI https://doi.org/10.2147/OTT.S136878
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Yao Dai
Zu-cheng Xie,1,* Yi-wu Dang,1,* Dan-ming Wei,1 Peng Chen,1 Rui-xue Tang,1 Qian Huang,1 Jiang-hua Liu,1,2,* Dian-zhong Luo1,*
1Department of Pathology, 2Department of Emergency Medicine, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, People’s Republic of China
*These authors contributed equally to this work
Purpose: Long noncoding RNAs (lncRNAs) are known to function as regulators in the development and occurrence of various tumors. MALAT1 is a highly conserved lncRNA and has vital functions in diverse tumors, including pancreatic cancer (PC). However, the underlying molecular regulatory mechanism involved in the occurrence and development of PC remains largely unknown. Thus, it is important to explore MALAT1 in PC and elucidate its function, which might offer a new perspective for clinical diagnosis and therapy.
Methods: First, we used the Gene Expression Omnibus, Oncomine, and The Cancer Genome Atlas databases to determine the clinical diagnostic and prognostic values of MALAT1. We next used our own GeneChip and The Cancer Genome Atlas database to collect the possible target genes of MALAT1 and further utilized a bioinformatics analysis to explore the underlying significant pathways that might be crucial in PC. Finally, we identified several key target genes of MALAT1 and hope to offer references for future research.
Results: We found that the expression of MALAT1 was significantly elevated in patients with PC. A receiver operating characteristics curve analysis showed a moderate diagnostic value (area under the curve =0.75, sensitivity =0.66, specificity =0.72). A total of 224 important overlapping genes were collected, and six hub genes (CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1) were identified, of which CCND1, MAPK8, and VEGFA, are important genes in PC. Several pathways, including the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway, were suggested to be the vital MALAT1 pathways in PC.
Conclusion: MALAT1 is suggested to be a promising diagnostic biomarker in PC. Six hub genes (CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1), and specifically CCND1, MAPK8, and VEGFA, might be key MALAT1 target genes in PC. Due to their possible clinical significance in PC, several pathways, such as the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway, are worthy of further study.
Keywords: MALAT1, pancreatic cancer, bioinformatics, target gene
Introduction
Pancreatic cancer (PC) is the seventh most common cause of cancer mortality and causes 330,000 deaths per year worldwide, accounting for approximately 4.0% of all cases of cancer.1 Currently, while the incidence of PC tends to linearly increase, the mortality of PC is high, and the prognosis is dismal. It is estimated that patients who present with early disease and have a negative resection margin only have a 5-year survival rate of 24%.2 There is an even higher mortality for unresectable patients. Currently, PC lacks a sensitive diagnosis marker to monitor the disease at an early stage and to predict the prognosis. Therefore, PC is in desperate need of a new index.
Long noncoding RNAs (lncRNAs) do not encode proteins and were once regarded as transcriptional noise or cloning artifacts.3,4 According to recent research, lncRNAs are thought to be regulators of cancer development, which make them a promising target for cancer treatment.5,6 MALAT1, with a length of over 8,000 nucleotides, is a highly conserved lncRNA that is derived from chromosome 11q13. It controls cancer cell proliferation, differentiation, apoptosis, and invasion by regulating the synthesis of proteins and the expression of genes. MALAT1 is well known as the first lncRNA, indicating the poor prognosis of non-small-cell lung cancer.7 Currently, emerging research has shown that MALAT1 plays a crucial role in ovarian cancer,8 esophageal cancer,9 prostate cancer10 breast cancer,11 thyroid cancer,12 nasopharyngeal carcinoma,13 and other types of malignant diseases. Previously, studies have found that elevated MALAT1 levels are involved in PC proliferation and metastasis through the stimulation of autophagy.14 Nevertheless, the potential molecular mechanism is still unclear and needs additional research. Over the last few years, there have been numerous novel techniques related to cancer mechanism and therapy exploration emerging. For instance, molecular docking and molecular dynamics simulation, which helps identify the cancer-associated single nucleotide polymorphisms and their possible molecular mechanism.15–17 And other analysis including protein–ligand interaction analysis, principal component analysis, shape analysis of binding pocket, and kinase inhibitor screening are also used to explore the therapeutic molecules for cancers.18,19 Researchers are also trying to figure out a methodology to optimize a synergistic and clinically achievable combination of multiple agents for cancer clinical therapy.20 Despite their particular perspectives in uncovering the cancer mechanism and therapy, the complicated and demanding operation of the experimental techniques themselves might limit their further development. As a result, we hope to use computational methods to explore the cancer mechanism by virtue of online databases. Thus, in this paper, we performed a comprehensive study to further explore the clinical significance and underlying molecular mechanism of MALAT1 in PC on a computational level.
To explore the relationship between MALAT1 and PC, we collected gene expression data and clinical data from patients using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). The relationship between MALAT1 and PC was further analyzed based on the TCGA and GEO data. For the purpose of exploring the molecular mechanism, we analyzed the overlapping genes in the differentially expressed genes after MALAT1 knockout and the coexpressed genes from TCGA. We performed a bioinformatics functional analysis for the overlapping genes and used Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Protein–Protein Interaction (PPI) databases.
Taken together, we hope to understand the clinical value of MALAT1 in PC and the molecular mechanism of MALAT1. We hope that the data shown here may offer new perspectives in future research and clinical application.
Materials and methods
Selection of microarrays in GEO datasets and profiles
For the purpose of understanding the diagnostic value of MALAT1 in PC, we systematically retrieved the GEO microarrays performed before December 2016 for a meta-analysis. The retrieval strategies were as follows: Pancrea* AND (adenocarcinoma OR carcinoma OR cancer OR neoplasm OR tumor OR tumour OR neoplas* OR malignan*). The inclusion criteria were as follows: 1) the enrolled subjects included patients with PC and normal control samples, 2) MALAT1 was expressed, detected, and available for extraction in the enrolled subjects, and 3) there were more than three subjects.
Validation of MALAT1 expression in PC based on the Oncomine database
The Oncomine database (http://www.oncomine.org) incorporates 264 independent datasets that include 35 cancer types and supports various methods of analysis, including molecular concepts analysis, interactome analysis, and meta-analysis.21 Thus, we adopted the Oncomine database to further validate the expression of MALAT1 in PC. The differential expression analysis was directly performed using Oncomine online analysis tools.
Identification of the clinical prognostic value of MALAT1 in PC
TCGA (http://cancergenome.nih.gov) data portal is one of the largest available public resources offering genomic, transcriptomic, methylomic, and copy number variation datasets for more than 20 cancer types.22,23 We evaluated the TCGA survival of patients with PC in two online analysis websites, cBioPortal for Cancer Genomics24,25 (http://www.cbioportal.org/) and OncoLnc26 (http://www.oncolnc.org/), which link to the TCGA database.
Identification of key differentially expressed genes associated with MALAT1 in PC
For the purpose of investigating the genes influenced by MALAT1, we performed GeneChip analysis after MALAT1 was knocked down. The human PC Miapaca-2 cell line was purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Wuhan, People’s Republic of China), and was maintained routinely in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA, USA). The four siRNAs for MALAT1 were designed on the internet (http://design.RNAi.jp/) and synthesized by Shanghai Jima company. Lentivirus MALAT1-shRNA was synthesized using lentivirus, packaged, and screened. The knockdown rate of MALAT1-stable cells was 76% in Miapaca-2 cell line with the best siRNA sequence (F:GGCAGCTTTAACAGATAACA; R:CCGTCCGACAAGGGTCATTCA).The three groups of control vector cells and MALAT1-shRNA stable cells were tested with GeneChip prime view human kit (Affymetrix., 901838, Santa Clara, CA, USA). Using MALAT1 knockdowns, we first acquired the differentially expressed genes. After performing GO analysis, we obtained the enriched GO data and the corresponding genes. Furthermore, we collected MALAT1 and mRNA gene coexpression data from the TCGA database. We adopted 0.15 as the cut-off value for screening the significant coexpression genes. Aiming at determining the most likely key differentially expressed MALAT1-associated genes in PC, we further correlated the enriched GO items with correlative genes and TCGA coexpression genes. The overlapping genes we obtained were used for further analysis.
Bioinformatics analysis of the overlapping genes
For the overlapping genes, The Database for Annotation, Visualization, and Integrated Discovery version 6.7 (https://david-d.ncifcrf.gov/), an online bioinformatics functional enrichment resource for a list of genes analysis,27,28 was used to perform GO enrichment and KEGG pathway analysis. The analysis was achieved via uploading the overlapping genes though the Functional Annotation porch. We chose those items whose P-values were below 0.05 for further study. Further, we used BiNGO, a plugin of Cytoscape 3.40,29 to visualize the GO enrichment pathway, which includes biological process (BP), cellular component (CC), and molecular function (MF). Meanwhile, we also aimed to identify the most crucial target genes of MALAT1 from the overlapping genes. The STRING database version 10 (http://string-db.org/) is aimed at providing a critical assessment and integration of PPI through direct (physical) and indirect (functional) associations network construction.30 Thus, STRING database was further used to perform a protein–protein interaction network analysis. The PPI network construction was achieved through StringApp, a plugin of Cytoscape 3.40, which links to STRING database. Interaction score (0.7) was adopted as a high confidence cut-off value to determine the PPI. Hub genes, which are the key genes in PC, were obtained from the network.
Statistical analysis
Stata 12.0 (Stata Corp., College Station, TX, USA) was used to perform a GEO microarray meta-analysis in this study. Receiver operating characteristic (ROC) curve analyses were utilized to evaluate the diagnostic value of MALAT1 in PC and normal tissue. Kaplan–Meier curves and the log-rank test were applied to analyze the effect of MALAT1 on the survival outcome of patients with PC. Only P<0.05 was considered statistically significant.
Results
Characteristics of the included datasets
A total of 27 eligible datasets published from 2008 to 2016 in the GEO database were used in this study. The characteristics of the selected datasets are shown in Table 1. In total, 764 PC samples and 469 healthy controls were used.
  | Table 1 Characteristics of the GEO datasets included in the meta-analysis |
Clinical diagnostic value of MALAT1 as a biomarker in PC based on the GEO datasets
After pooling the data from the 27 eligible datasets, we found that there was significant heterogeneity among the datasets (I2=61.1%, P=0.000, Figure 1). As a result, we performed a sensitivity analysis to screen the possible datasets that might cause the heterogeneity. As can be seen in Figure 2, two datasets, namely, GSE15471 and GSE62165, may contribute to significant heterogeneity. Thereby, we removed the two datasets and recalculated the pooled standard mean difference. As shown in Figure 3, the heterogeneity appears to have disappeared (I2=44.9%, P=0.009). The pooled standard mean difference was 0.23 (95% confidence interval [CI]: 0.10–0.37, P=0.001), which indicates a significant difference in the effect of MALAT1 between patients with PC and healthy control patients. The expression of MALAT1 was significantly upregulated. Publication bias among the 25 datasets was assessed through Begg’s funnel plot. As shown in Figure 4, the funnel plot was basically considered symmetrical. Taking P>0.05 as cut-off value, we found that P=0.414 was obtained by Begg’s test, which indicated that no significant publication bias existed. Furthermore, a summary ROC curve revealed that the area under the curve was 0.75 (95% CI: 0.71–0.78), and the overall sensitivity and specificity was 0.66 (95% CI: 0.57–0.74) and 0.72 (95% CI: 0.61–0.81) (Figures 5 and 6A). The pooled diagnostic odds ratio was also calculated as 4.41 (95% CI: 2.71–7.17), which indicates that upregulated MALAT1 results in a higher risk of PC (Figure 6B).
  | Figure 4 Begg’s funnel plot evaluating the publication bias among the 25 datasets. |
MALAT1 expression difference validation based on the Oncomine database
Five PC datasets in the Oncomine database were adopted for the validation of MALAT1 expression (Grutzmann’s dataset [http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-950]; Badea’s datasets [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15471]; lacobuzio-Donahue’s dataset [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3654; http://genome-www.stanford.edu/pancreatic1/index.shtml]; Pie’s dataset [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16515]; Ishikawa’s dataset [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1542]). In Grutzmann’s and Badea’s datasets, the expression of MALAT1 was significantly elevated in PC, which was in accordance with the results of the meta-analysis (Figure 7A and B). The P-values for these datasets were 0.025 and <0.01, respectively. However, the remaining three datasets did not show a statistically significant difference in MALAT1 expression (Figure 7C–E).
  | Figure 7 Box plot validating the expression of MALAT1 in the PC group and the healthy control group based on the Oncomine database. |
Prognostic value of MALAT1 based on the TCGA database
For the analysis using the cBioPortal for Cancer Genomics, the survival data for a total of 178 PC patients was analyzed. We found 10 of 178 patients had alterations in the expression of MALAT1 (Supplementary material 1). Thus, we evaluated the survival of the cases with MALAT1 alteration compared to those without MALAT1 alteration. The log-rank test showed that the P-value was 0.0380 (Figure 8A). As reported on the Oncolnc website, we analyzed the survival of patients with high MALAT1 expression and compared it to that of patients with low MALAT1 expression in a total of 174 patients with PC. It showed a trend that high MALAT1 expression indicates better survival, although no statistical significance was observed (P=0.304, Figure 8B).
Collection of the overlapping genes
After knocking down MALAT1, we performed a GO enrichment analysis and collected 656 enriched GO gene symbols. Furthermore, using 0.15 as P-value cut-off, we obtained 5,887 coexpressed genes from the TCGA database. We then intersected the two part genes and gathered 224 overlapping genes (Supplementary material 2), which might become the crucial targets genes of MALAT1 in PC.
Bioinformatics analysis of the overlapping genes
As displayed in Table 2, the overlapping genes contain three enriched GO pathway categories: BP, CC, and MF. In the BP category, the enriched items were mainly intracellular transport, regulation of phosphorus metabolic process, regulation of phosphate process, etc. In the CC category, membrane-enclosed lumen, nuclear lumen, and nuclear chromosome, among others, were the main components of the enrichment. In regards to the MF category, protein dimerization activity, nucleoside binding, and purine nucleoside binding ranked as the top three enriched items. All of the enriched GO pathways, including the BP, CC, and MF categories, were further visualized through Cytoscape 3.40 and the results are displayed in Supplementary materials 3 and 4 and Figure 9, respectively. In the KEGG pathway analysis, we identified the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway as the top three enriched pathways (Table 2). Interestingly, we also found that the PC pathway was one of the significantly enriched pathways. Five target genes (VEGFC, CCND1, PIK3CB, VEGFA, and MAPK8) were involved in the PC pathway. A PPI was constructed (Supplementary material 5) and six hub genes (CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1) were identified (Figure 10).
  | Table 2 Significant GO and KEGG items of the overlapping genes |
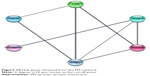  | Figure 10 Hub genes obtained from the PPI network. |
Discussion
In this paper, we first discovered that the expression of MALAT1 was significantly upregulated in PC patients by using a meta-analysis of 27 published GEO microarrays, which included a total of 764 patients with PC and 469 healthy controls. Further validation via the Oncomine database was consistent with the GEO microarray meta-analysis. Using the TCGA database, we acquired the survival data of the patients and investigated the influence of the alteration of MALAT1 expression on the patients’ prognosis. Furthermore, using our own GeneChip and TCGA database, we collected and intersected the enriched GO genes after knocking down MALAT1 and the coexpressed genes in TCGA. Further bioinformatics analysis of the overlapping genes, which aims to explore the underlying molecular regulatory mechanism, was performed.
During the past few years, alteration of MALAT1 has been found to be involved in malignancies and other diseases. Shuai et al31 performed a meta-analysis that revealed that MALAT1 served as a promising biomarker for the prognosis of numerous malignancies, including digestive system cancers, urinary system cancers, and respiratory system cancers. In our study, we observed upregulated MALAT1, which is consistent with other previous reports, such as those by Li et al,14 Pang et al,32 and Liu et al.33 Elevated MALAT1 has also been reported in gastric cancer,34 hepatocellular carcinoma,35 gallbladder cancer,36 esophageal cancer,37 and clear cell renal cell carcinoma.38 Considering all of the above information, MALAT1 appears to play a distinct role in different tumors, which provides a brand new perspective for future cancer therapy.
In the present study, we discovered that MALAT1 showed a moderate performance in diagnosing PC after meta-analysis of a large number of PC, and corresponding control, samples. The area under the curve for the summary ROC curve was 0.75, with a sensitivity of 0.66 and a specificity of 0.72. Thus far in the published literature, there is no report evaluating the diagnostic value of MALAT1 in PC through comprehensive meta-analysis. As a consequence, the information in this study will provide a valuable reference for clinical PC diagnosis. To date, several studies have reported that higher MALAT1 expression suggests poorer survival.31,32 Regarding our results, we found that patients with MALAT1 alteration had better survival than patients without MALAT1 alteration (P=0.0380). We also found that the high expression of MALAT1 suggested better survival in the Kaplan–Meier curves. However, the P-values did not show statistical significance. Therefore, the exact prognostic value of MALAT1 in PC still remains unknown.
Since lncRNAs exert their regulatory function by specifically binding with target genes, we also aimed to determine possible targets of MALAT1 and to further explore the possible molecular pathways. Through a GO enrichment analysis, some top-ranked GO items, such as intracellular transport, membrane-enclosed lumen, and protein dimerization activity, were found to be possible crucial events in PC development. Regarding the KEGG pathway analysis, several significant pathways, such as the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway, were discovered. Importantly, the PC pathway was also one of the enriched KEGG pathways. Five genes, VEGFC, CCND1, PIK3CB, VEGFA, and MAPK8, are target genes of MALAT1, which might also be key target genes in PC. Interestingly, according to previous studies, we found that the mTOR signaling pathway has been reported to be involved in PC.39,40 Additionally, the MAPK signaling pathway also has been reported to play a role in PC.41 Therefore, to some extent, the pathways identified here might provide more evidence for elucidating the potential molecular mechanism in PC. Finally, the PPI network suggests that six hub genes (CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1) might serve as pivotal target genes of MALAT1 in PC. We discovered that CCND1, MAPK8, and VEGFA also participate in the PC pathway, as mentioned. As a consequence, we concentrate on the top three genes (CCND1, MAPK8, and VEGFA), which may be important genes, for detailed discussion.
CCND1, also known as BCL1, encodes a highly conserved cyclin protein, which exerts regulatory functions on CDK kinases. Mutation, amplification, and overexpression of CCND1 alter cell cycle progression, which might contribute to tumorigenesis. As a result, CCND1 alteration was frequently discovered in various tumors. The CCND1 genotype has been reported to be related to the risk for various cancers, such as colorectal cancer,42 acute lymphoblastic leukemia,43 glioma,44 esophageal squamous cell carcinoma,45 gastric cancer,46 and nasopharyngeal carcinoma.47,48 Moreover, CCND1 was also reported to play different roles in cancer survival, such as in lung adenocarcinoma,49,50 prostate cancer,51 meningioma52 gastric adenocarcinoma,53 and colorectal cancer.54 In regards to PC, Deharvengt et al55 reported that CCND1 is a target of a B lentivirus-delivered shRNA, which exerts suppressive effects on the growth, invasiveness, tumorigenicity, and proangiogenic potential of PC. Another study found that CCND1 was a strong prognostic biomarker for PC survival.56 Taken together, it is obvious that CCND1 is involved in various cancers, including PC. However, whether it is related to MALAT1 has not been reported to date, and further studies are required.
MAPK8, also named JNK or JNK1, belongs to the MAPK kinase family that acts as an integration point in numerous cellular processes, such as proliferation, differentiation, transcription regulation, and development. Increasing amounts of evidence have proven that MAPK8 has a crucial role in a wide variety of cancers. Chang et al57 discovered that JNK1 activation was capable of predicting poor prognosis in hepatocellular carcinoma. Moreover, Okada et al58 reported that JNK can inhibit temozolomide, thus acting as a rational therapeutic biomarker in glioblastoma. JNK activity has also been reported in endometrial cancer,59 melanoma,60 colon cancer,61 etc. In regard to PC, Sahu et al62 reported that JNK was involved in benzyl isothiocyanate (BITC)-mediated G(2)/M arrest to mediate apoptosis in human PC cells. Another study also found that JNK1 is activated by Irofulven treatment, which induces apoptosis in human PC cells.63 Therefore, based on the abovementioned information, MAPK8 could possibly serve as a target gene of MALAT1 in PC, but additional studies are needed.
VEGFA, also known as VPF, VEGF, and MVCD1, is a member of the PDGF/VEGF growth factor family. Encoding a heparin-binding protein, it induces the proliferation and migration of vascular endothelial cells, which is fundamental for physiological and pathological angiogenesis. Upregulated VEGFA is common in many tumors and is related to tumor stage and progression. Liu et al64 reported that VEGFA polymorphisms might play a potential role in the development and clinical outcome of hepatocellular carcinoma. VEGFA has also been found to be inhibited by miR-1, which exerts inhibitory functions in osteosarcoma cells, as well as promotes proliferation, migration, and invasion.65 Furthermore, Zeng et al66 found that downregulation of VEGFA can inhibit proliferation, migration, and invasion and can promote apoptosis in renal clear cell carcinoma. VEGFA in tumors has also been studied in breast cancer,67,68 laryngeal carcinoma,69 colorectal cancer,70,71 bladder cancer,72 and lung cancer.73 In PC, Liu et al74 found that VEGFA was involved in the Twist/miR-497/VEGFA axis, which is significantly correlated to metastasis and angiogenesis in PC. VEGFA secretion has also been reported to be regulated by myoferlin and affects tumor-associated angiogenesis in PC.75 In summary, as VEGFA has been shown to be important in cancers, it is suggested that VEGFA is a target of MALAT1 in PC. However, more studies are needed.
There are limitations in this study. Here, we have attempted to explore the relationship between MALAT1 and PC clinical parameters based on the TCGA database. However, no statistical significance was observed. Therefore, it still remains to be seen if there is a correlation between MALAT1 and PC. In addition, in this study, the exploration occurred at a bioinformatics level. Further experiments are needed to validate these assumptions.
In sum, we discovered that upregulated MALAT1 acts as a biomarker for the diagnosis of PC. The alteration or upregulation of MALAT1 expression tends to predict better prognostic outcome in PC. Several pathways, such as the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway, might be targeted by MALAT1 and participate in the PC process. Six hub genes (CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1), and specifically CCND1, MAPK8 and VEGFA, were identified as key target genes of MALAT1 in PC. Here, we provided clinical reference values and underlying molecular mechanisms for MALAT1 in PC. These findings still need to be validated in future studies.
Conclusion
MALAT1 seems to act as a promising diagnostic biomarker in PC. Six hub genes (CCND1, MAPK8, VEGFA, FOS, CDH1, and HSP90AA1), specifically CCND1, MAPK8, and VEGFA, might be key correlative genes of MALAT1 in PC. Several pathways related to MALAT1, such as the mTOR signaling pathway, pathways in cancer, and the MAPK signaling pathway may play a crucial role in PC. The information we have obtained might shed light on clinical application and future research. Nevertheless, more experimental studies are needed to further validate these findings.
Acknowledgments
The study was supported by the Fund of National Natural Science Foundation of China (NSFC 81560448), the Natural Science Foundation of Guangxi, China (2014GXNSFBA118167), and Guangxi Medical University Students Innovative Project (201610598100). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Lei Q, Zheng H, Bi J, et al. Whole grain intake reduces pancreatic cancer risk: a meta-analysis of observational studies. Medicine (Baltimore). 2016;95(9):e2747. | ||
Ambe CM, Mahipal A, Fulp J, Chen L, Malafa MP. Effect of metformin use on survival in resectable pancreatic cancer: a single-institution experience and review of the literature. PLoS One. 2016;11(3):e0151632. | ||
Been JV, Mackenbach JP, Millett C, Basu S, Sheikh A. Tobacco control policies and perinatal and child health: a systematic review and meta-analysis protocol. BMJ Open. 2015;5(9):e008398. | ||
Shi X, Sun M, Wu Y, et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol. 2015;36(2):503–513. | ||
Yuan J, Yue H, Zhang M, et al. Transcriptional profiling analysis and functional prediction of long noncoding RNAs in cancer. Oncotarget. 2016;7(7):8131–8142. | ||
Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. | ||
Wei Y, Niu B. Role of MALAT1 as a prognostic factor for survival in various cancers: a systematic review of the literature with meta-analysis. Dis Markers. 2015;2015:164635. | ||
Lei R, Xue M, Zhang L, Lin Z. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther. 2017;10:35–46. | ||
Li Z, Zhou Y, Tu B, Bu Y, Liu A, Xie C. Long noncoding RNA MALAT1 affects the efficacy of radiotherapy for esophageal squamous cell carcinoma by regulating Cks1 expression. J Oral Pathol Med. Epub 2016 Dec 09. | ||
Aiello A, Bacci L, Re A, et al. MALAT1 and HOTAIR long non-coding rnas play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci Rep. 2016;6:38414. | ||
Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7(26):40418–40436. | ||
Huang JK, Ma L, Song WH, et al. MALAT1 promotes the proliferation and invasion of thyroid cancer cells via regulating the expression of IQGAP1. Biomed Pharmacother. 2016;83:1–7. | ||
Hua WF, Zhong Q, Xia TL, et al. RBM24 suppresses cancer progression by upregulating miR-25 to target MALAT1 in nasopharyngeal carcinoma. Cell Death Dis. 2016;7(9):e2352. | ||
Li L, Chen H, Gao Y, et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. 2016;15(9):2232–2243. | ||
Kamaraj B, Rajendran V, Sethumadhavan R, Purohit R. In-silico screening of cancer associated mutation on PLK1 protein and its structural consequences. J Mol Model. 2013;19(12):5587–5599. | ||
Kumar A, Purohit R. Use of long term molecular dynamics simulation in predicting cancer associated SNPs. PLoS Comput Biol. 2014;10(4):e1003318. | ||
Rajendran V. Structural analysis of oncogenic mutation of isocitrate dehydrogenase 1. Mol biosyst. 2016;12(7):2276–2287. | ||
Rajendran V, Gopalakrishnan C, Purohit R. Impact of point mutation P29S in RAC1 on tumorigenesis. Tumour Biol. 2016;37(11):15293–15304. | ||
Baranski Z, Booij TH, Kuijjer ML, et al. MEK inhibition induces apoptosis in osteosarcoma cells with constitutive ERK1/2 phosphorylation. Genes Cancer. 2015;6(11–12):503–512. | ||
Yu D, Kahen E, Cubitt CL, et al. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci Rep. 2015;5:16991. | ||
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. | ||
Deng M, Bragelmann J, Schultze JL, Perner S. Web-TCGA: an online platform for integrated analysis of molecular cancer data sets. BMC Bioinformatics. 2016;17:72. | ||
Krasnov GS, Dmitriev AA, Melnikova NV, et al. CrossHub: a tool for multi-way analysis of The Cancer Genome Atlas (TCGA) in the context of gene expression regulation mechanisms. Nucleic Acids Res. 2016;44(7):e62. | ||
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. | ||
Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. | ||
Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J Computer Science. 2016;2:e67. | ||
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. | ||
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. | ||
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. | ||
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. | ||
Shuai P, Zhou Y, Gong B, et al. Long noncoding RNA MALAT1 can serve as a valuable biomarker for prognosis and lymph node metastasis in various cancers: a meta-analysis. SpringerPlus. 2016;5(1):1721. | ||
Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36(4):2403–2407. | ||
Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15(7):2971–2977. | ||
Lee NK, Lee JH, Ivan C, et al. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer. 2017;17(1):46. | ||
Malakar P, Shilo A, Mogilavsky A, et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 up-regulation and mTOR activation. Cancer Res. 2017;77(5):1155–1167. | ||
Wang SH, Zhang WJ, Wu XC, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7(25):37857–37867. | ||
Huang C, Yu Z, Yang H, Lin Y. Increased MALAT1 expression predicts poor prognosis in esophageal cancer patients. Biomed Pharmacother. 2016;83:8–13. | ||
Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36(4):2947–2955. | ||
He L, Wu Y, Lin L, et al. Hispidulin, a small flavonoid molecule, suppresses the angiogenesis and growth of human pancreatic cancer by targeting vascular endothelial growth factor receptor 2-mediated PI3K/Akt/mTOR signaling pathway. Cancer Sci. 2011;102(1):219–225. | ||
Kagawa S, Takano S, Yoshitomi H, et al. Akt/mTOR signaling pathway is crucial for gemcitabine resistance induced by Annexin II in pancreatic cancer cells. J Surg Res. 2012;178(2):758–767. | ||
Wu YY, Ma TL, Ge ZJ, et al. JWA gene regulates PANC-1 pancreatic cancer cell behaviors through MEK-ERK1/2 of the MAPK signaling pathway. Oncol Lett. 2014;8(4):1859–1863. | ||
Huang CY, Tsai CW, Hsu CM, Chang WS, Shui HA, Bau DT. The significant association of CCND1 genotypes with colorectal cancer in Taiwan. Tumour Biol. 2015;36(8):6533–6540. | ||
Xue Y, Rong L, Tong N, Wang M, Zhang Z, Fang Y. CCND1 G870A polymorphism is associated with toxicity of methotrexate in childhood acute lymphoblastic leukemia. Int J Clin Exp Pathol. 2015;8(9):11594–11600. | ||
Zong H, Cao L, Ma C, et al. Association between the G870A polymorphism of Cyclin D1 gene and glioma risk. Tumour Biol. 2014;35(8):8095–8101. | ||
Hussain S, M Y, Thakur N, et al. Association of cyclin D1 gene polymorphisms with risk of esophageal squamous cell carcinoma in Kashmir Valley: a high risk area. Mol Carcinog. 2011;50(7):487–498. | ||
Kuo HW, Huang CY, Fu CK, et al. The significant association of CCND1 genotypes with gastric cancer in Taiwan. Anticancer Res. 2014;34(9):4963–4968. | ||
Shih LC, Tsai CW, Tsai MH, et al. Association of cyclin D1 genotypes with nasopharyngeal carcinoma risk. Anticancer Res. 2012;32(3):1093–1098. | ||
Liao D, Wu Y, Pu X, et al. Cyclin D1 G870A polymorphism and risk of nasopharyngeal carcinoma: a case-control study and meta-analysis. PLoS One. 2014;9(11):e113299. | ||
Xu P, Zhao M, Liu Z, et al. Elevated nuclear CCND1 expression confers an unfavorable prognosis for early stage lung adenocarcinoma patients. Int J Clin Exp Pathol. 2015;8(12):15887–15894. | ||
Lee E, Jin D, Lee BB, et al. Negative effect of cyclin D1 overexpression on recurrence-free survival in stage II-IIIA lung adenocarcinoma and its expression modulation by vorinostat in vitro. BMC Cancer. 2015;15:982. | ||
Tsai H, Morais CL, Alshalalfa M, et al. Cyclin D1 loss distinguishes prostatic small-cell carcinoma from most prostatic adenocarcinomas. Clin Cancer Res. 2015;21(24):5619–5629. | ||
Cheng G, Zhang L, Lv W, Dong C, Wang Y, Zhang J. Overexpression of cyclin D1 in meningioma is associated with malignancy grade and causes abnormalities in apoptosis, invasion and cell cycle progression. Medical Oncol. 2015;32(1):439. | ||
Ma L, Wang X, Lan F, et al. Prognostic value of differential CCND1 expression in patients with resected gastric adenocarcinoma. Medical Oncol. 2015;32(1):338. | ||
Li Y, Wei J, Xu C, Zhao Z, You T. Prognostic significance of cyclin D1 expression in colorectal cancer: a meta-analysis of observational studies. PLoS One. 2014;9(4):e94508. | ||
Deharvengt SJ, Gunn JR, Pickett SB, Korc M. Intratumoral delivery of shRNA targeting cyclin D1 attenuates pancreatic cancer growth. Cancer Gene Ther. 2010;17(5):325–333. | ||
Bachmann K, Neumann A, Hinsch A, et al. Cyclin D1 is a strong prognostic factor for survival in pancreatic cancer: analysis of CD G870A polymorphism, FISH and immunohistochemistry. J Surg Oncol. 2015;111(3):316–323. | ||
Chang Q, Chen J, Beezhold KJ, Castranova V, Shi X, Chen F. JNK1 activation predicts the prognostic outcome of the human hepatocellular carcinoma. Mol Cancer. 2009;8:64. | ||
Okada M, Sato A, Shibuya K, et al. JNK contributes to temozolomide resistance of stem-like glioblastoma cells via regulation of MGMT expression. Int J Oncol. 2014;44(2):591–599. | ||
Reno EM, Haughian JM, Jackson TA, Thorne AM, Bradford AP. c-Jun N-terminal kinase regulates apoptosis in endometrial cancer cells. Apoptosis. 2009;14(6):809–820. | ||
Alexaki VI, Javelaud D, Mauviel A. JNK supports survival in melanoma cells by controlling cell cycle arrest and apoptosis. Pigment Cell Melanoma Res. 2008;21(4):429–438. | ||
Mahalingam D, Keane M, Pirianov G, Mehmet H, Samali A, Szegezdi E. Differential activation of JNK1 isoforms by TRAIL receptors modulate apoptosis of colon cancer cell lines. Br J Cancer. 2009;100(9):1415–1424. | ||
Sahu RP, Zhang R, Batra S, Shi Y, Srivastava SK. Benzyl isothiocyanate-mediated generation of reactive oxygen species causes cell cycle arrest and induces apoptosis via activation of MAPK in human pancreatic cancer cells. Carcinogenesis. 2009;30(10):1744–1753. | ||
Wang W, Waters SJ, MacDonald JR, et al. Irofulven (6-hydroxymethylacylfulvene, MGI 114)-induced apoptosis in human pancreatic cancer cells is mediated by ERK and JNK kinases. Anticancer Res. 2002;22(2A):559–564. | ||
Liu F, Luo L, Wei Y, et al. Association of VEGFA polymorphisms with susceptibility and clinical outcome of hepatocellular carcinoma in a Chinese han population. Oncotarget. 2017;8(10):16488–16497. | ||
Niu J, Sun Y, Guo Q, Niu D, Liu B. miR-1 Inhibits cell growth, migration, and invasion by targeting VEGFA in osteosarcoma cells. Dis Markers. 2016;2016:7068986. | ||
Zeng FC, Zeng MQ, Huang L, et al. Downregulation of VEGFA inhibits proliferation, promotes apoptosis, and suppresses migration and invasion of renal clear cell carcinoma. Onco Targets Ther. 2016;9:2131–2141. | ||
Vieira-Monteiro Hde A, Freitas-Alves DR, Sobral-Leite M, et al. Prognostic evaluation of VEGFA genotypes and haplotypes in a cohort of Brazilian women with non metastatic breast cancer. Cancer Biol Ther. 2016;17(6):674–683. | ||
Wu J, Yuan P, Mao Q, et al. miR-613 inhibits proliferation and invasion of breast cancer cell via VEGFA. Biochem Biophys Res Commun. 2016;478(1):274–278. | ||
Xu L, Shen B, Chen T, Dong P. miR-203 is involved in the laryngeal carcinoma pathogenesis via targeting VEGFA and Cox-2. Onco Targets Ther. 2016;9:4629–4637. | ||
Zhang W, Zou C, Pan L, et al. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37(3):1123–1133. | ||
Takahashi N, Iwasa S, Taniguchi H, et al. Prognostic role of ERBB2, MET and VEGFA expression in metastatic colorectal cancer patients treated with anti-EGFR antibodies. Br J Cancer. 2016;114(9):1003–1011. | ||
Gao Y, Wu K, Chen Y, et al. Beyond proliferation: KLF5 promotes angiogenesis of bladder cancer through directly regulating VEGFA transcription. Oncotarget. 2015;6(41):43791–43805. | ||
Zhang SD, McCrudden CM, Kwok HF. Prognostic significance of combining VEGFA, FLT1 and KDR mRNA expression in lung cancer. Oncol Lett. 2015;10(3):1893–1901. | ||
Liu A, Huang C, Cai X, Xu J, Yang D. Twist promotes angiogenesis in pancreatic cancer by targeting miR-497/VEGFA axis. Oncotarget. 2016;7(18):25801–25814. | ||
Fahmy K, Gonzalez A, Arafa M, et al. Myoferlin plays a key role in VEGFA secretion and impacts tumor-associated angiogenesis in human pancreas cancer. Int J Cancer. 2016;138(3):652–663. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.