Back to Journals » International Journal of General Medicine » Volume 16
Clinical Characterization of the Expression of Insulin-Like Growth Factor Binding Protein 1 and Tumor Immunosuppression Caused by Ferroptosis of Neutrophils in Non-Small Cell Lung Cancer
Authors Wang Y , Xing L, Deng L, Wang X, Xu D, Wang B , Zhang Z
Received 10 January 2023
Accepted for publication 28 February 2023
Published 21 March 2023 Volume 2023:16 Pages 997—1015
DOI https://doi.org/10.2147/IJGM.S401225
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Video abstract presented by Yuandi Wang.
Views: 94
Yuandi Wang,1,2,* Lijuan Xing,2,* Lexiu Deng,1,2 Xinsheng Wang,2 Dandan Xu,2 Bu Wang,2 Zhihua Zhang2,*
1Graduate School, Hebei North University, Zhangjiakou City, Hebei Province, People’s Republic of China; 2Department of Respiratory and Critical Care Clinical Medicine, The First Affiliated Hospital of Hebei North University, Zhangjiakou City, Hebei Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhihua Zhang, Department of Respiratory and Critical Care Clinical Medicine, The First Affiliated Hospital of Hebei North University, Zhangjiakou City, Hebei Province, People’s Republic of China, Tel +86 0313 8033598, Email [email protected]
Purpose: The efficacy of immunotherapy for non-small cell lung cancer (NSCLC) is limited owing to cold tumors and drug resistance. Therefore, it is important to identify the molecular mechanisms underlying immune evasion in NSCLC. Spontaneous ferroptosis of neutrophils has been suggested as a key mechanism of immunosuppression in cancer. Insulin-like growth factor binding protein 1 (IGFBP1) plays an important role in immune infiltration in several cancers. However, the role of IGFBP1 in NSCLC is unknown. Therefore, in this study, we aimed to investigate the association of IGFBP1 mRNA expression with infiltration of myeloid-derived suppressor cells and prognosis in NSCLC.
Patients and Methods: Retrospective RNA-seq data from 990 patients in the Cancer Genome Atlas (TCGA) database were analyzed in relation to patient clinical characteristics. The Timer2 database was used to assess immune infiltration, and the FerrDb V2 database was used to obtain ferroptosis-related genes. Finally, the results were validated by the proteomic analysis of serum samples collected from six patients with NSCLC and six healthy individuals.
Results: IGFBP1 expression was enriched in lung adenocarcinoma samples and positively correlated with the pathological grade of NSCLC. IGFBP1 expression was an independent prognostic factor for the overall survival of patients with NSCLC. In addition, IGFBP1 expression correlated with myeloid-derived suppressor cell infiltration. Notably, Gene Ontology analysis of IGFBP1-related genes revealed that the major molecular functions of their protein products were related to NADP+ 1-oxidoreductase activity. Furthermore, expression levels of multiple ferroptosis suppressor genes positively correlated with IGFBP1 expression.
Conclusion: High IGFBP1 expression indicates a poor prognosis in patients with NSCLC, which may be related to tumor immunosuppression caused by neutrophil ferroptosis.
Keywords: immune infiltration, immunotherapy, lung adenocarcinoma, lung squamous cell carcinoma, myeloid-derived suppressor cells
Introduction
Primary lung tumors are among the most common causes of fatal cancer in adults.1 According to the revised 2021 World Health Organization classification of lung tumors, non-small cell lung cancer (NSCLC) accounts for approximately 85% of all diagnosed lung cancers. Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the most common histological types of lung cancer.2 NSCLC is resistant to all current standard treatment measures, including surgery, radiotherapy, and systemic chemotherapy, and most patients with NSCLC eventually relapse.3 Therefore, new radical treatment strategies are urgently needed for NSCLC.
Immunotherapy has shown substantial efficacy in the treatment of many tumors.4 To date, the most remarkable advancement in cancer therapy is the reactivation of T cell immune responses to tumors following the inhibition of the immune checkpoint activity.5 Immunotherapy has been used with considerable success in melanoma, renal cell carcinoma, and Hodgkin lymphoma. However, only 20% of patients with NSCLC benefit from immunotherapy.6 In addition, a recent single-cell RNA sequencing analysis showed that in addition to T lymphocytes, natural killer (NK) cells and different functional subtypes of macrophages and neutrophils are important players in the heterogeneous tumor immune microenvironment.7 Therefore, the mechanism of immunosuppression in NSCLC requires further investigation.
Ferroptosis, an iron-dependent mechanism of regulated cell death triggered by the accumulation of products of toxic lipid peroxidation, is a promising therapeutic target in cancer.8 Although ferroptosis inducers are highly effective in killing tumor cells in vitro, no obvious effect has been observed in experimental animal models in vivo, except in immunodeficient mice.9,10 A recent study has shown that spontaneous ferroptosis of neutrophils in the tumor microenvironment promoted the release of oxygenated lipids and inhibited the activity of T cells, mediating a strong immunosuppressive effect.11 Therefore, these findings suggest that ferroptosis is closely associated with immunosuppression in tumors.
Insulin-like growth factor binding protein 1 (IGFBP1) circulates in the plasma and binds insulin-like growth factors (IGF) I and II, extending their half-life and altering their interaction with cell surface receptors. In addition to its important roles in cell migration and metabolism,12 IGFBP1 regulates tumor immune infiltration in clear cell renal cell carcinoma,13 esophageal cancer,14 and metastatic melanoma.15 However, the role of IGFBP1 in NSCLC is unknown. Therefore, the role and mechanism of action of IGFBP1 in NSCLC-associated cellular immunosuppression need to be investigated as part of the wider search for new therapeutic targets that could be used to immunize cold tumors and improve the efficacy of lung cancer immunotherapy.
In this study, we aimed to examine the association between IGFBP1 expression and NSCLC prognosis. We analyzed IGFBP1 mRNA expression profile in LUAD and LUSC and investigated the correlation between IGFBP1 expression and clinicopathological characteristics and survival of patients with NSCLC using data from The Cancer Genome Atlas (TCGA). We further analyzed the relationship between IGFBP1 and molecules involved in immune infiltration and ferroptosis. Finally, serum proteome profiles of patients with lung cancer and healthy individuals were analyzed to detect differences in expression levels of IGFBP1 and its related proteins. Our results provide a basis for the design of future immunotherapies targeting IGFBP1 for the treatment of NSCLC.
Materials and Methods
Patients and Blood Collection
TCGA public database data (https://tcgadata.nci.nih.gov/tcga/tcgaDownload.jsp) were downloaded to analyze clinical features of patients with NSCLC and RNA sequencing data. Data from 1091 patients were obtained (LUAD, N = 586; LUSC, N = 505). Of these, samples of 14 individuals with LUAD and 5 individuals with LUSC were excluded because their tissue sequencing samples were pathological wax blocks. In addition, samples of two patients were excluded because they had recurrent LUAD, and samples of another 80 patients were also excluded because of missing data on IGFBP1 expression. Thus, samples of 990 patients with NSCLC were included in this study.
Six patients with NSCLC, pathologically diagnosed with LUAD or LUSC, were recruited from the First Affiliated Hospital of Hebei North University for validation. None of the patients had received chemotherapy or immunotherapy. The control group consisted of six healthy individuals from the physical examination center who had no history of malignant tumors or acute infection. Peripheral blood samples were collected from fasting patients who agreed to be included in the study, stored at room temperature for 2 h, centrifuged at 3000 g for 10 min, and then, serum samples were collected and frozen at −80 °C. This study complied with the Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of the Hebei North University (No.: K2022071). All participants signed an informed consent form. This study was approved by the Chinese Clinical Trial Registry (No.: ChiCTR2200065162).
Identification of the Expression of Mutated Genes Affecting IGFBP1 Expression
To identify mutations in genes affecting IGFBP1 gene expression, the “Target” section of the online muTarget tool (https://www.mutarget.com/) was used.16 This tool, developed by the Department of Bioinformatics of the Semmelweis University (Budapest, Hungary), links mutational status to changes in gene expression in solid tumors. A procedure was set up to screen somatic mutations with the mutation rate of at least 2% that were shown to be associated with IGFBP1 expression changes. The cutoff P-value was 0.01. The cutoff fold change value (FC) was 1.44.
Promoter Methylation
The epigenetic regulation of IGFBP1 expression was determined by investigating IGFBP1 promoter methylation using The University of Alabama at Birmingham Cancer data analysis portal (UALCAN).17,18 All promoter methylation data for LUAD and LUSC were collected from the TCGA dataset. The database was created in 2017 and updated in March 2022.
Functional Enrichment Analysis
The list of featured genes or cell clusters most associated with IGFBP1 in the training set was uploaded to the Database for Annotation, Visualization, and Integrated Discovery (DAVID; Dec. 2021). Official gene symbols were selected as identifiers, and Homo sapiens was selected as the species. Finally, the top six results of the gene ontology (GO) analysis were sorted according to P-values that were below 0.05. Protein interaction analysis was performed using the STRING database.19
Immune Infiltration
To investigate the relationship between IGFBP1 expression level and immune infiltration, the TIMER2.0 database (http://timer.comp-genomics.org/) was used.21 The cutoff value of the correlation coefficient R was 0.2, and the cutoff P-value was 0.05. The TISIDB database was used to perform immune cell classification and verify the relationship between IGFBP1 expression and infiltration of immune cells.
Characteristic Molecules and Ferroptosis
Suppressor genes encoding proteins that prevent ferroptosis have been obtained from FerrDb V2.20 The cutoff correlation coefficient R value for IGFBP1-related genes was 0.2, and the cutoff P-value was 0.05. The Venn online portal was used to process the data of intersection genes.The Kaplan-Meier plotter (kmplot.com) was used to investigate the association between IGFBP1-related ferroptosis molecules in NSCLC prognosis.
Gene Effect Score
The gene effect score of IGFBP1 was obtained from UALCAN data according to CRISPR knockout screening published by Project Achilles at the Broad Institute and Project Score at the Wellcome Sanger Institute.22 Negative scores indicated growth inhibition and/or cell death after gene knockdown. Scores were standardized, and the median score for the non-essential genes was 0, whereas that for independently identified common essential genes was −1.
Isolation and Identification of Serum Proteins
Proteomic profiling was performed using the serum of patients with NSCLC and healthy controls. For protein extraction and quantitative quality control, proteins were extracted using High Select™ HSA/Immunoglobulin Depletion Mini Spin Columns (Thermo Fisher Scientific). Proteins were then quantified using a microplate reader (H4MFPTAD; BioTek, Winooski, VT, USA). The total amount of the detected protein was derived from the standard curve, and the corresponding concentration was calculated. SDS-PAGE and Coomassie Brilliant Blue staining were used to evaluate protein degradation and distribution of proteins with different molecular weights.
For the enzymatic hydrolysis of the proteins, the protein solutions of the 12 studied individuals were pretreated, trypsin was added according to the protein-enzyme ratio of 50:1, and the proteins were digested at 37 °C for 12–16 h.
For liquid chromatography-mass spectrometry, enzymatically digested samples were separated using a nanoliter flow-rate HPLC liquid system. The column was equilibrated with 95% liquid A. Samples were loaded from an autosampler to a mass spectrometry pre-column and then separated using an analytical column. Each sample was separated by capillary HPLC and analyzed by mass spectrometry using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
For protein identification, raw mass spectrometry data were screened, and label-free quantitative analysis was performed using the Uniprot-proteome UP000005640.fasta database. Database files were analyzed using Proteome Discoverer 2.4 software.
Screening of Differentially Expressed Proteins in Serum Samples
A serum protein was considered downregulated if FC ≤ 0.667 and upregulated if FC ≥1.5. A protein was considered to have unchanged expression if 0.667 < FC < 1.5. Hierarchical clustering was used to analyze the similarities between the samples. Protein expression level FC values in the two groups were compared by the t-test, and the difference was considered statistically significant if P < 0.05. A volcano plot was drawn after log transformation of FC and the test statistic P-values were used to visualize significant differences between the two groups of samples.
Statistical Analysis
Statistical analysis and visualization were performed using R (version 4.1.1) and GraphPad Prism (version 8.0.2). The Mann–Whitney U-test was used to compare significant differences between the two groups. The nonparametric statistical analysis of multiple independent samples was used to compare multiple groups. The Spearman correlation analysis was used to analyze significant correlations between the two groups. The Kaplan–Meier curve was used to evaluate the prognostic value of IGFBP1 expression. The significance of prognostic values was determined using the Log rank test. Effects were considered statistically significant if P < 0.05.
Results
Clinicopathological Characteristics of Patients with Different IGFBP1 Expression Levels
We investigated the association between patient characteristics and IGFBP1 expression and found that patients with different IGFBP1 expression levels had distinct clinicopathological features. IGFBP1 mRNA expression levels, pathological grades, and pathological types were asymmetrically distributed in the TCGA dataset (Figure 1A). Stratified comparative analysis showed no significant difference in IGFBP1 expression between paracancerous tissues (control) and samples from all patients with lung cancer, between samples from individuals with different ethnic background, or between healthy individuals and those with LUAD (Figure 1B and C; S1). However, IGBFP1 expression levels in the LUSC group were significantly lower than those in the control and LUAD groups (P = 0.008 and P < 0.001, respectively; Figure 1D and E). No significant difference in IGFBP1 expression was observed between the two sexes (Figure 1F). However, IGFBP1 expression in the death group was significantly higher than that in the survival group (P = 0.0470). In addition, the expression of IGFBP1 increased (P = 0.039) with the increase in the pathological grade, showing a gradual change between the groups. Collectively, these results indicate that IGFBP1 is differentially expressed in individuals with different pathological types of NSCLC and that IGFBP1 expression is upregulated in LUAD compared to its level in LUSC.
Effects of Different Gene Mutations and Promoter Methylation Status on the Epigenetic Regulation of IGFBP1 Expression
Gene mutations associated with changes in IGFBP1 expression were obtained from the muTarget database. In LUAD, mutations in 30 genes were related to changes in IGFBP1 expression (Figure 2A), among which mutations in 11 genes were associated with upregulated IGFBP1 expression compared to that in samples without such mutations. The top three genes in this category were KCNQ2 (FC = 13.08), RNF165 (FC = 5.51), and KEAP1 (FC = 4.55). Additionally, mutations in 19 genes, including ALDH1L1, EGFR, TRIML2, ZNF99, NHS, CHSY3, LAX1, and IL2R, were associated with significantly downregulated (by approximately 98%) IGFBP1 expression compared to that in samples without mutations in those genes. In LUSC, mutations in 38 genes were associated with altered IGFBP1 expression (Figure 2B). Among these, mutations in 22 genes were associated with upregulated IGFBP1 expression, and the top 3 genes were UNC45B (FC = 115.26), RPH3A (FC = 100.95), and DCLK1 (FC = 92.3). Additionally, mutations in 16 genes including NPY2R, ST5, NCOA1, and CD1D, were associated with downregulated (by approximately 95%) IGFBP1 expression in LUSC compared to that in samples without such mutations. Compared with its level in healthy individuals, IGFBP1 expression in patients with LUAD (71%) and LUSC (66%) was significantly downregulated by NFE2L2 mutations.
Promoter methylation data were obtained from UALCAN to investigate the epigenetic regulation of IGFBP1 expression in NSCLC. The median methylation level of the IGFBP1 promoter in LUAD was 0.472 (0.438–0.500) Beta, which was lower than that in normal tissues (0.502; range: 0.495–0.518, P<0.001; Figure 2C). There was no significant difference in the methylation level of the IGFBP1 promoter between samples from LUAD pathological stage 3 and those healthy groups (P > 0.05). However, the methylation level of IGFBP1 in LUAD pathological stages 1, 2, and 4 was lower than that in the control group (P < 0.05; Figure 2D). The degree of IGFBP1 methylation in LUSC was similar to that in LUAD. Moreover, the median methylation level of the IGFBP1 promoter in LUSC was 0.446 (0.403–0.491) Beta, which was lower than that in healthy tissues (0.501; range: 0.486–0.513; P <0.001; Figure 2E). There was no significant difference in the methylation level of the IGFBP1 promoter between LUSC pathological stage 4 and control groups (P > 0.05), whereas the level of IGFBP1 methylation in LUSC pathological stage 1–3 groups was lower than that in the control group (P < 0.05; Figure 2F). These results suggest that common gene mutations affect IGFBP1 expression and that IGFBP1 methylation is significantly downregulated in NSCLC. High levels of methylation are associated with gene expression silencing. Changes in the levels of this epigenetic mark could elevate IGFBP1 expression in cancer tissues compared with normal tissues. Furthermore, decreased methylation could also elevate IGFBP1 expression proportionally to the pathological grade of NSCLC.
IGFBP1 mRNA Promotes Cell Proliferation in Tumor Cells, and Its High Expression is Associated with Poor Prognosis
To explore IGFBP1 role in NSCLC, biological process (BPs), cellular localization (CCs) and molecular function (MFs) of genes that were found to affect IGFBP1 expression were examined. Genes related to IGFBP1 were screened using the Pearson correlation analysis (r > 0.2, P < 0.05), and GO analysis was performed on this set of genes. In the TCGA data, the top six entries with the largest number of enriched genes were screened and ranked using the corrected P-value. The BPs significantly associated with IGFBP1 included drug response, negative regulation of endopeptidase activity, protein processing, response to lipopolysaccharide, cell-to-cell signaling, and positive regulation of cell proliferation (Figure 3A). In addition, the most relevant CCs of IGFBP1 were the extracellular space and extracellular vesicles (Figure 3B). MFs associated with these genes were d-aldolase 1-dehydrogenase activity, NADP+ 1-oxidoreductase activity, and protein binding (Figure 3C).
In addition, we investigated IGFBP1 protein interactions using the String database. IGFBP1 was closely associated with IGF1, IGF2, and FOXO family proteins (Figure 3D). We then used survival analysis to describe the association between IGFBP1 expression data and overall survival in 991 NSCLC patients from the TCGA database (Figure 4A). IGFBP1 expression level was considered high (IGFBP1high) if it was > 75% of the IGFBP1 expression and low (IGFBP1low), if it was below the IGFBP1high level. There were 249 patients with IGFBP1high and 742 with IGFBP1low in our analysis. The overall survival of patients with IGFBP1high was shorter than that of patients with IGFBP1low (P < 0.001; Figure 4A). To validate the results of TCGA survival assessment, we used RNA sequencing data from a larger cohort of 1925 patients with NSCLC in the online Kaplan–Meier tool (www.kmplot.com) (Figure 4B). This cohort had 956 and 969 patients with high and low levels of IGFBP1 expression, respectively. The relationship between IGFBP1 expression and survival according to the Kaplan-Meier online tool analysis was similar to that obtained from the analysis of the TCGA dataset: NSCLC patients with IGFBP1high had shorter overall survival than patients with IGFBP1low (P < 0.001).
Different IGFBP1 Expression Levels and Degree of Immune Infiltration in Distinct Pathological Types of NSCLC
Genes related to IGFBP1 expression were obtained from the Timer2 database. In LUAD, the extent of bone marrow-derived suppressor cell (MDSC) infiltration positively correlated with IGFBP1 expression (r = −0.205, P <0.001) (Figure 5A). IGFBP1 expression negatively correlated with the numbers of mast cells (r = −0.308, P = <0.001), NK T cells (r = −0.208, P<0.001), and B cells (r = −0.214, P =<0.001) (Figure 5B–D). There was no correlation between LUSC and degrees of infiltration of MDSCs, mast cells, T cells, NK cells, and B cells, except for a weak correlation with the number of endothelial cells (r = 0.105, P =0.0222; Figure 5E–I). In addition, the expression of the FATP2 (SLC27A2) gene, encoding a transporter that positively regulates ferroptosis in tumor neutrophils and polymorphonuclear (PMN)-MDSCs, positively correlated with IGFBP1 expression in NSCLC cells (r = 0.17, P =<0.001; Figure 5J). In conclusion, IGFBP1 expression in NSCLC cells positively correlated with the extent of MDSC infiltration in LUAD and with the expression of SLC27A2, which encodes a positive regulator of ferroptosis in PMN-MDSCs.
IGFBP1-Associated Ferroptosis Suppressor Genes are Associated with Poor Prognosis
Given the strong link between immunosuppression and ferroptosis, we explored the relationship between expression levels of IGFBP1 and genes encoding ferroptosis inhibitors. The Pearson correlation analysis showed that genes related to IGFBP1 (r > 0.2, P < 0.05) overlapped with 195 genes encoding ferroptosis inhibitors in the FerrDb V2 database, of which seven were related to IGFBP1 expression: AKR1C1, AKR1C2, AKR1C3, GCLC, FURIN, VDAC2, and SLC7A11 (Figure 6A). Therefore, the relationship between these intersecting genes and tumor prognosis was determined using the Kaplan–Meier survival analysis. In the Kaplan-Meier online tool, the median of the population was used as the cut-off value, and the patients were divided into groups with high and low expression levels of each of the seven intersection genes. For AKR1C2, AKR1C3, and GCLC (Figure 6B–D), there was no significant difference in overall survival risk between patients with high and low expression levels (P>0.05). In contrast, high expression levels of AKR1C1 (hazard ratio [HR] = 1.161; 95% confidence interval [CI]: 1.02–1.32; P = 0.02), SLCTA 11 (HR = 1.15; 95% CI: 1.02–1.31; P = 0.027), and VDAC2 (HR = 1.66; 95% CI: 1.46–1.89; P<0.001) (Figure 6E–G) were associated with poor prognosis in patients with NSCLC. The relationship between the expression level of FURIN and overall survival was nominally similar (HR = 1.12, 95% CI: 0.99–1.127), although the result did not pass the set criterion for statistical significance (P = 0.08); therefore, the prognostic potential of FURIN expression for NSCLC remains uncertain (Figure 6H).
IGFBP1 Downregulation Inhibits Tumor Cell Growth and IGFBP1 Protein is Highly Expressed in the Blood of Patients with NSCLC
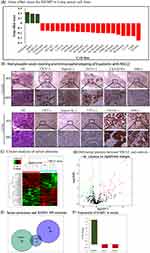
A total of 23 cell lines with IGFBP1 gene effect score ≥ 0.15 were obtained from the UALCAN database (Figure 7A). In the NCIH2887 (0.2292), NCIH2126 (0.1942), and SHP77 (0.1904) cell lines, IGFBP1 knockdown had a slight growth-promoting effect. In the 20 remaining cell lines, IGFBP1 knockdown resulted in cell death or growth inhibition, with the largest such effect seen in the NCIH23 cell line (−0.3524).
To validate our results, six serum samples from patients with NSCLC (mean age 69.83 ± 3.80 years) and six samples from control individuals (mean age 49.50 ± 6.70 years) were collected for mass spectrometry. The samples from NSCLC cases were diagnosed as LUAD (N = 3) and LUSC (N = 3) using hematoxylin-eosin (H&E) staining and immunophenotyping before serum mass spectrometry analysis (Figure 7B). H&E staining showed that the nuclei of the lesion area were dark stained with hematoxylin and their shapes were irregular. Immunohistochemistry staining showed that LUAD samples were CK7 (+), Napsin (+), TFF1 (+), CK5/6 (+/−), and P40 (−). In contrast, LUSC carcinomas were CK7 (−), Napsin (−), TFF1 (−), CK5/6 (+), and P40 (+). The label-free quantitative proteomic analysis was used to compare proteins in the serum of these 12 individuals (Table 1). The proteins were effectively separated in the molecular weight range of 25–180 kDa without protein degradation, and the overall protein pattern had a similar band shape (Figure S2). In total, 5700 unique peptides and 516 proteins were identified. Hierarchical clustering analysis revealed that 83 proteins were significantly differentially expressed between the NSCLC and control groups (Figure 7C), of which 44 were upregulated and 39 were downregulated (Figure 7D). Filaggrin A (FC = 34.11615, P < 0.001) was the most upregulated protein (Table 2), and fumarase acetate esterase (FC = 0.229015, P = 0.015) was the most downregulated protein in NSCLC (Table 3). The eight proteins identified in the protein-protein interaction analysis of IGFBP1 were compared to the set of the differentially expressed proteins identified in the serum of patients, and common proteins were revealed (Figure 7E). IGFBP1 (FC = 3.814) was upregulated, whereas IGF2 (FC = −0.56) and AHSG (FC = −0.58) were downregulated in the serum of patients with NSCLC compared to their levels in the serum of healthy individuals (Figure 7F). In conclusion, IGFBP1 knockdown mostly negatively affected the growth of LUAD cells. Furthermore, the IGFBP1 protein was found to be abundant in the serum of patients with NSCLC.
 |
Table 1 Subject Characteristics and Protein Extraction Parameters |
 |
Table 2 Differentially Expressed Upregulated Proteins in Non-Small Cell Lung Cancer Samples |
 |
Table 3 Differentially Expressed Downregulated Proteins in Non-Small Cell Lung Cancer Samples |
Discussion
NSCLC is the most common and lethal cancer of the respiratory system.23 Conventional therapies, which usually include surgical resection, radiotherapy, and chemotherapy, fail to show satisfactory results in patients with NSCLC.24 Additionally, the clinical efficacy of immunotherapy in patients with NSCLC is limited,25,26 owing to the unique suppressive immune microenvironment; however, the mechanism of this phenomenon remains to be established.7,11 Therefore, elucidation of the molecular mechanisms underlying tumor-induced immunosuppression and the development of novel therapies that target these mechanisms are essential for the effective treatment of NSCLC.
We found that post-transcriptional methylation of IGFPB1 likely increased its expression in cancer tissues with disease progression. In LUAD and LUSC, when NFE2L2 was mutated, IGFBP1 expression was significantly lower than that in samples from healthy individuals. GO analysis indicated that IGFBP1 is involved in the positive regulation of intercellular signaling and cell proliferation and may affect the activity of D-threo-aldose 1 and NADP+ 1-oxidoreductase. Survival analysis showed that upregulated expression of IGFBP1 predicted poor prognosis in NSCLC. IGFBP1 may be associated with infiltration of neutrophils. Expression of SLC27A2, encoding a fatty acid transporter that promotes ferroptosis of PMN-MDSCs, positively correlated with the overall expression level of IGFBP1 in NSCLC. At the same time, expression levels of mRNAs encoding ferroptosis inhibitors AKR1C1, SLCTA11, and VDAC2 also positively correlated with IGFBP expression, and high expression levels of those genes predicted poor prognosis of NSCLC. IGFBP1 knockout resulted in cell death or growth inhibition of the majority of lung cancer lines tested. Finally, using serum protein profiling, we verified that in patients with NSCLC, IGFBP1 expression level was higher, whereas expression levels of AHSG,\andIGF2were lower compared to the respective levels in healthy individuals. Overall, our results suggest thatIGFBP1 expression in NSCLC, is associated with tumor immunosuppression caused by ferroptosis of neutrophils.
IGFBP1 expression in NSCLC involves a complex regulatory mechanism and is closely related to oxidative metabolic reactions. Although the expression of IGFBP1 in NSCLC is not significantly higher than that in normal control tissues, post-transcriptional epigenetic modifications may affect the expression of IGFBP1 protein. Methylation is a common mode of epigenetic modification, and generally hypermethylated water is thought to be associated with gene silencing.27 We further found that the promoter of IGFPB1 was hypomethylated in cancer tissues and hypermethylated in normal tissues. This epigenetic change likely increases IGFBP1 expression in cancer tissues in a disease progression-dependent manner. In NSCLC, distinct tumor gene expression profiles may partly account for the differences in clinical manifestations, biological characteristics, diagnosis, and treatment of LUAD and LUSC.2,28 In our study, we showed that IGFBP1 expression in LUAD was higher than that in LUSC. Notably, compared with its level in healthy population, IGFBP1 expression in both LUAD and LUSC was significantly downregulated in the presence of NFE2L2 mutations. NFE2L2 is a major regulator of oxidative stress signaling, and limits oxidative damage during ferroptosis through the trans-activation of several cellular protective genes associated with iron metabolism, glutathione metabolism (including SLC7A11), and reactive oxygen detoxification enzymes (including AKR1C1–3).29 At present, the link between the important defense mechanism of NFE2L2 mutation against ferroptosis and IGFBP1 downregulation is not clear. According to the GO analysis, IGFBP1 can affect activities of d-aldolase 1-dehydrogenase and NADP+ 1-oxidoreductase, regulating oxidation reactions. Ethnicity is an important factor affecting tumor gene expression profiles; herein, we found no significant effect of ethnicity on IGFBP1 expression in our study.30
During tumor development, some tumor cells may escape antitumor immunity.31 Changes in the characteristic tumor microenvironment, such as tumor necrosis and hypoxia, promote the accumulation of neutrophils in the adjacent tissues, which profoundly affects tumor progression.32 Much attention has been paid to the role of neutrophils in tumor immunity.11,33 Some studies showing that neutrophils and granulocyte colony-stimulating factor promote tumor development34,35 and others pointing out that the effector function of neutrophils is related to antitumor immune suppression.11,36 Recent studies have found that the neutrophil precursors PMN-MDSCs are vulnerable to ferroptosis which promotes the suppression of tumor immune responses.11 The present study suggests that IGFBP1 expression correlates with MDSC infiltration in LUAD, However, no such correlation was observed in LUSC. Because the effect of the transcriptional modification on protein activity is important,27 it remains to be tested whether IGFBP1 expression is altered in various subtypes of NSCLC. An earlier study showed that the neutrophil-derived factor azurocidin shows protease activity that cleaves several IGFBP proteins, including IGFBP1, which is detected at higher levels in inflammatory states.37 Inflammatory infiltration is also an important feature of the tumor microenvironment,38 hence it is possible that neutrophils upregulated IGFBP1 expression in the tumor microenvironment. A study involving a large sample of 666 patients confirmed that IGFBP1 is a marker gene for poor prognosis in NSCLC associated with neutrophils and hypoxic microenvironment.39 In our study, IGFBP1 transcript levels increased with pathological grade, and IGFBP1 overexpression was detected by comparing the protein expression profiles of NSCLC patients and healthy subjects. IGFBP1, a characteristic gene related to energy metabolism in lung adenocarcinoma,40 is overexpressed in the serum of individuals with NSCLC.41 In addition, it has been shown that peripheral B cells inhibit B cell regeneration in aging through the TNF-α/IGFBP-1/IGF-1 immuno-endocrine axis. In our study, IGFBP1 expression was also found to correlate positively with the abundance of infiltrated B cells.42 In summary, IGFBP1 was highly expressed in the immune microenvironment of NSCLC, where it may be regulated by central granulocytes, and correlated with the level of PMN-MDSC infiltration.
Much attention has been paid to the relationship between ferroptosis and suppression of antitumor immunity. Cancer cells with initial ferroptosis decrease DC maturation are poorly phagocytosed, and inhibit antigen cross-presentation.43 Secondly, ferroptotic damage of immune cells such as PMN-MDSCs in the tumor microenvironment promotes immune escape.11 IGFBP1 regulates tumor immune responses in renal clear cell carcinoma,13 esophageal cancer,14 and metastatic melanoma.15
GO evidence suggests that signaling pathways such as IGFBP1/IGF1 may regulate NADP+ 1-oxidoreductase activity in ferroptosis. This conjecture is supported by evidence that ferroptosis is accompanied by GPX4 depletion and increased NADP+/NADPH ratio, inhibiting the conversion of cystine to cysteine and synthesis of GSH.8–10 Moreover, genomic and proteomic studies of the human adaptive response to high altitude/hypoxia environment also support IGFBP1 involvement in key molecular functions such as IGF-1 signaling, ferroptosis, iron homeostasis, and cell cycle regulation.44 We further analyzed the inhibitors of ferroptosis associated with IGFBP1 and found that expression levels of many of them were associated with poor prognosis in NSCLC. A recent report also confirmed the importance of IGFBP1 in lung cancer, as risk scores based on gene signatures of IGFBP1 and other related secreted proteins correlated with tumor prognosis as well as resistance to antitumor immunity in lung adenocarcinoma. There is currently no evidence linking IGFBP1 with ferroptosis in lung cancer cells, but early studies have shown that IGFBP1 protein can be efficiently cleaved by enzymes secreted by immune cells, such as central granulocyte-derived proteases cathepsin G and elastase, in vitro and in vivo.34 Tumor hypoxia and inflammatory infiltration provide strong support for the aggregation and activation of central granulocytes.38 Our preliminary study also showed that LUAD is associated with the infiltration of PMN-MDSCs with ferroptosis fragility. In conclusion, the activity of IGFBP1 in NSCLC is regulated by neutrophils, which, in turn, affects oxidoreductases, such as NADP+ 1-oxidoreductases, and ferroptosis of PMN-MDSCs.
This study has some limitations.Because of the lack of material limitation, we were unable to verify the expression of IGFBP1 in serum and further analyze in situ immune cells profiles and protein expression. Although we identified IGFBP1 as a therapeutic target, we did not investigate new IGFBP1 inhibitors. Therefore, whether targeting of IGFBP1 can effectively treat NSCLC remains to be established. In addition, we evaluated the effect of IGFBP1 knockdown expression on NSCLC cell line growth only in a single experiment. We aim to validate these results in an animal model of NSCLC, which will undoubtedly improve the precision of our conclusions.
Conclusion
In conclusion, in this study, we revealed that high IGFBP1 expression is associated with poor prognosis. This relationship may be explained by the role of IGFBP1 in ferroptosis of neutrophils, which, in turn, may mediate cancer immunosuppression. We suggest that IGFBP1 may be used as a prognostic indicator and a therapeutic target in NSCLC.
Acknowledgments
This study was funded by a Grant for Scientific Research Projects on Traditional Chinese Medicine in 2023 (ID 2023097) by the Hebei Provincial Administration of Traditional Chinese Medicine and the Hebei Province Graduate Innovation Funding Project (ID CXZZSS2022150). We thank Shanghai Origingene Bio-Pharm Technology Co., Ltd. (Shanghai, China) for their contribution to library construction, sequencing, and data analysis.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. 2022;17(3):362–387. doi:10.1016/j.jtho.2021.11.003
3. Deb D, Moore AC, Roy UB. The 2021 global lung cancer therapy landscape. J Thorac Oncol. 2022;17(7):931–936. doi:10.1016/j.jtho.2022.03.018
4. Pala L, Sala I, Oriecuia C, et al. Association of anticancer immune checkpoint inhibitors with patient-reported outcomes assessed in randomized clinical trials: a systematic review and meta-analysis. JAMA Netw Open. 2022;5(8):e2226252. doi:10.1001/jamanetworkopen.2022.26252
5. Xiong A, Wang J, Zhou C. Immunotherapy in the first-line treatment of NSCLC: current status and future directions in China. Front Oncol. 2021;11:757993. doi:10.3389/fonc.2021.757993
6. Bote H, Mesas A, Baena J, Herrera M, Paz-Ares L. Emerging immune checkpoint inhibitors for the treatment of non-small cell lung cancer. Expert Opin Emerg Drugs. 2022;27(3):289–300. doi:10.1080/14728214.2022.2113377
7. Wang C, Yu Q, Song T, et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduct Target Ther. 2022;7(1):289. doi:10.1038/s41392-022-01130-8
8. Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22(7):381–396. doi:10.1038/s41568-022-00459-0
9. Dierge E, Debock E, Guilbaud C, et al. Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell Metab. 2021;33(8):1701–1715.e5. doi:10.1016/j.cmet.2021.05.016
10. Zou Y, Henry WS, Ricq EL, et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature. 2020;585(7826):603–608. doi:10.1038/s41586-020-2732-8
11. Kim R, Hashimoto A, Markosyan N, et al. Ferroptosis of tumour neutrophils causes immune suppression in cancer. Nature. 2022;612(7939):338–346. doi:10.1038/s41586-022-05443-0
12. Ye H, Adane B, Khan N, et al. Subversion of systemic glucose metabolism as a mechanism to support the growth of leukemia cells. Cancer Cell. 2018;34(4):659–673.e6. doi:10.1016/j.ccell.2018.08.016
13. Xu T, Gao S, Liu J, Huang Y, Chen K, Zhang X. MMP9 and IGFBP1 regulate tumor immune and drive tumor progression in clear cell renal cell carcinoma. J Cancer. 2021;12(8):2243–2257. doi:10.7150/jca.48664
14. Liu M, Yan W, Chen D, et al. hiWNT3Alo subtype in esophageal cancer predicts response and prolonged survival with PD-(L)1 inhibitor. Biology. 2022;11(11):1575. doi:10.3390/biology11111575
15. Li Y, Gao Y, Chu W, Lv J, Li Z, Shi T. Differences in and verification of genetic alterations in chemotherapy and immunotherapy for metastatic melanoma. Aging. 2021;13(20):23672–23688. doi:10.18632/aging.203640
16. Nagy Á, Győrffy B. muTarget: a platform linking gene expression changes and mutation status in solid tumors. Int J Cancer. 2021;148(2):502–511. doi:10.1002/ijc.33283
17. Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi:10.1016/j.neo.2022.01.001
18. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
19. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi:10.1093/nar/gkaa1074
20. Zhou N, Yuan X, Du Q, et al. FerrDb V2: update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 2023;51(D1):D571–D582. doi:10.1093/nar/gkac935
21. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
22. Dempster JM, Boyle I, Vazquez F, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22(1):343. doi:10.1186/s13059-021-02540-7
23. Brouwer AF, Engle JM, Jeon J, Meza R. Sociodemographic survival disparities for lung cancer in the United States, 2000–2016. J Natl Cancer Inst. 2022;114(11):1492–1500. doi:10.1093/jnci/djac144
24. Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun. 2022;42(10):937–970. doi:10.1002/cac2.12359
25. Ramos-Esquivel A. Immunotherapy in non-small-cell lung cancer: new data support its use, but some challenges remain. Thorax. 2022;77(12):1159–1160. doi:10.1136/thorax-2022-219472
26. Kashefizadeh A, Kazemizadeh H. Immunogenic cell death (ICD)-inducers in non-small-cell lung carcinoma (NSCLC): current knowledge and future perspective. Clin Transl Oncol. 2022;25(2):316–322. doi:10.1007/s12094-022-02949-x
27. Oliva M, Demanelis K, Lu Y, et al. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat Genet. 2023;55(1):112–122. doi:10.1038/s41588-022-01248-z
28. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
29. Hellyer JA, Padda SK, Diehn M, Wakelee HA. Clinical Implications of KEAP1-NFE2L2 Mutations in NSCLC. J Thorac Oncol. 2021;16(3):395–403. doi:10.1016/j.jtho.2020.11.015
30. Ma F, Laster K, Dong Z. The comparison of cancer gene mutation frequencies in Chinese and U.S. patient populations. Nat Commun. 2022;13(1):5651. doi:10.1038/s41467-022-33351-4
31. Cascone T, Fradette J, Pradhan M, Gibbons DL. Tumor immunology and immunotherapy of non-small-cell lung cancer. Cold Spring Harb Perspect Med. 2022;12(5):a037895. doi:10.1101/cshperspect.a037895
32. Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, Mantovani A. Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer. 2020;20(9):485–503. doi:10.1038/s41568-020-0281-y
33. Salcher S, Sturm G, Horvath L, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022;40(12):1503–1520.e8. doi:10.1016/j.ccell.2022.10.008
34. Zahid KR, Raza U, Tumbath S, Jiang L, Xu W, Huang X. Neutrophils: musketeers against immunotherapy. Front Oncol. 2022;12:975981. doi:10.3389/fonc.2022.975981
35. Teijeira A, Garasa S, Ochoa MC, et al. IL8, neutrophils, and NETs in a collusion against cancer immunity and immunotherapy. Clin Cancer Res. 2021;27(9):2383–2393. doi:10.1158/1078-0432.CCR-20-1319
36. Cui C, Chakraborty K, Tang XA, et al. Neutrophil elastase selectively kills cancer cells and attenuates tumorigenesis. Cell. 2021;184(12):3163–3177.e21. doi:10.1016/j.cell.2021.04.016
37. Brandt K, Lundell K, Brismar K. Neutrophil-derived azurocidin cleaves insulin-like growth factor-binding protein-1, −2 and −4. Growth Horm IGF Res. 2011;21(3):167–173. doi:10.1016/j.ghir.2011.04.003
38. Mair F, Erickson JR, Frutoso M, et al. Extricating human tumour immune alterations from tissue inflammation. Nature. 2022;605(7911):728–735. doi:10.1038/s41586-022-04718-w
39. Zhang C, Tang B, Hu J, et al. Neutrophils correlate with hypoxia microenvironment and promote progression of non-small-cell lung cancer. Bioengineered. 2021;12(1):8872–8884. doi:10.1080/21655979.2021.1987820
40. Mu T, Li H, Li X. Prognostic implication of energy metabolism-related gene signatures in lung adenocarcinoma. Front Oncol. 2022;12:867470. doi:10.3389/fonc.2022.867470
41. Li F, Cao Y, Li J, et al. The clinical significance of serum adipocytokines level in patients with lung cancer. J Thorac Dis. 2019;11(8):3547–3555. doi:10.21037/jtd.2019.07.66
42. Dowery R, Benhamou D, Benchetrit E, et al. Peripheral B cells repress B-cell regeneration in aging through a TNF-α/IGFBP-1/IGF-1 immune-endocrine axis. Blood. 2021;138(19):1817–1829. doi:10.1182/blood.2021012428
43. Wiernicki B, Maschalidi S, Pinney J, et al. Cancer cells dying from ferroptosis impede dendritic cell-mediated anti-tumor immunity. Nat Commun. 2022;13(1):3676. doi:10.1038/s41467-022-31218-2
44. Sharma V, Varshney R, Sethy NK. Human adaptation to high altitude: a review of convergence between genomic and proteomic signatures. Hum Genomics. 2022;16(1):21. doi:10.1186/s40246-022-00395-y
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.