Back to Journals » Infection and Drug Resistance » Volume 13
Clinical and Microbiological Prognostic Factors of in-Hospital Mortality Caused by Hypervirulent Klebsiella pneumoniae Infections: A Retrospective Study in a Tertiary Hospital in Southwestern China
Authors Tang Y, Liu H, Zhao J, Yi M, Yuan Y, Xia Y
Received 12 August 2020
Accepted for publication 26 September 2020
Published 21 October 2020 Volume 2020:13 Pages 3739—3749
DOI https://doi.org/10.2147/IDR.S276642
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Yu Tang, Hang Liu, Jinxin Zhao, Miao Yi, Yaling Yuan, Yun Xia
Department of Clinical Laboratory, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Yun Xia
Department of Clinical Laboratory, The First Affiliated Hospital of Chongqing Medical University, No. 1 Youyi Road, Yuzhong District, Chongqing 400016, People’s Republic of China
Tel +86-23-89012742
Fax +86-23-89012513
Email [email protected]
Purpose: Hypervirulent klebsiella pneumoniae (hvKP) is responsible for various invasive diseases and associated with high mortality. However, the clinical and microbiological factors of hvKP infection that influence prognosis have not been well studied. The purpose of this study was to evaluate the prognostic factors for in-hospital mortality of patients with hvKP infections, mainly focusing on clinical and microbiological characteristics.
Methods: A retrospective study was conducted in hvKP strains which positive for iucA and string test. According to the clinical outcomes during hospitalization, hvKP-infected patients were divided into non-survivor and survivor groups. The clinical characteristics, capsule types, multi-locus sequence types (MLST), virulence genes and antimicrobial susceptibility were compared between those of the two groups.
Results: A total of 135 patients were demonstrated to be with hvKP infections, with a prevalence rate of 22% among all the klebsiella pneumoniae infected cases. Sixteen of these patients died during hospitalization, with an in-hospital mortality rate of 11.9%. Univariate analysis confirmed that admission to the intensive care unit (ICU) (p=0.008), antimicrobial resistance of hvKP to ampicillin/sulbactam (p=0.028), cefepime (p=0.033), aztreonam (p=0.049) and harboring iroN gene (p=0.023) were associated with in-hospital mortality. On the contrary, the rmpA gene showed an inverse association with in-hospital mortality (p=0.017). Multivariate logistic regression analysis revealed that ICU admission (odds ratio [OR]=3.452, 95% confidence interval [CI]=1.052– 11.329; P=0.041) and iroN carriage (OR=9.278, 95% CI=1.654– 52.035; P=0.011) were independent prognostic factors for the in-hospital mortality of patients with hvKP infections.
Conclusion: Emerging hvKP infection may lead to relatively high in-hospital mortality. ICU admission and iroN carriage were independent prognostic factors for the in-hospital mortality of patients with hvKP infections.
Keywords: hypervirulent Klebsiella pneumoniae, prognostic factors, iucA, iroN, in-hospital mortality
Introduction
As an opportunistic pathogen, Klebsiella pneumoniae is responsible for various infections such as pneumonia, intra-abdominal infection, urinary tract infection and bacteremia.1 The elderly and immunocompromised populations are susceptible to the classic Klebsiella pneumoniae (cKP).2 Unlike cKP, the emerging hypervirulent Klebsiella pneumonia (hvKP) can cause community-acquired infections in patients without major comorbidities or even young healthy individuals.3 Of note, the hvKP strain, generally considered as normal intestinal flora, has the ability to invade distal organs and tissues of the host, resulting in irreversible severe damage and higher clinical mortality.4 However, the reported death-rate of hvKP-infected patients fluctuates greatly in different countries and regions: 17% in Spain, and up to 41.2% in China, the huge difference may at least partially be caused by the misclassification of cKP and hvKP.5,6 So, the effective isolation and accurate classification of these strains are of great significance. However, the identification standard of hvKP is imperfect and controversial. Though initially defined as a hypermucoviscous phenotype, it is now more inclined to be termed as strain carrying a series of virulence genes. However, completely ignoring their high-mucoid characteristic is infeasible. After all, numerous virulence genes have been demonstrated to be associated with bacterial mucoid and capsule phenotype. The increasing capsule leads to increased anti-phagocytosis of bacteria, attenuated early inflammatory response in host and enhanced antagonistic complement-mediated effects.7–9 Recently, the combination of hypermucoviscous phenotype and aerobactin genotype has been suggested as an effective way to differentiate cKP and hvKP.10,11 The aerobactin, as one of the siderophores secreted by hvKP, has long been appreciated as an important factor to enhance the virulence and cytotoxicity of hvKP.12
Currently, extended-spectrum–lactamases (ESBLs) and carbapenemases-producing hvKP strains deserve particular attention owing to the difficulty of treatment, which have been proposed to be potential prognostic factors for mortality.11,13 The ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) strains are formed through the acquisition of a pLVPK-like plasmid by the ST11 carbapenem-resistant cKP strains, and are especially worrisome due to their fatal outcomes.14 Except for the virulent characteristics of the bacteria itself elucidating partial cause of death, a mounting of latent factors also affect the patients’ clinical outcomes, among which comorbidities and immunological status are well recognized as important host-associated ones. For example, in previous studies of risk factors for infection, patients with comorbidities of diabetes and malignancy are more vulnerable to hvKP infection.11 Diabetics have impaired neutrophil responses and phagocytic capabilities, while the amounts of immune cells decreased due to the side effects of cytotoxic therapies in patients with malignant tumors.15,16 A common feature of both comorbidities is that they lead to a decline in the host’s ability to defend against bacteria. The deficiency of immune defense ability combined with other factors such as increased pathogenicity and antimicrobial resistance of bacteria may lead to an increase in deaths in patients with hvKP infections.
So far, the research on the prevalence and in-hospital mortality rates of patients with hvKP infections in China is limited, and there is still a lack of systematic research on the prognostic factors influencing the in-hospital mortality of patients with hvKP infections. Therefore, we conducted a retrospective analysis of the prognostic factors for patients with hvKP infections, mainly focusing on the clinical characteristics of the patients and microbiological characteristics of the strains including capsule types, multi-locus sequence types (MLST), virulence genes and antimicrobial resistance.
Methods
Study Design and Definition
A retrospective study was conducted from February 2018 to June 2019 in the First Affiliated Hospital of Chongqing Medical University, a 3200-bed tertiary teaching hospital. Confirmation of Klebsiella pneumoniae (KP) isolates was performed by VITEK MS system or VITEK 2 compact system (BioMérieux, Marcy-l’Étoile, France). Firstly, a total of 613 cases positive for KP culture were collected consecutively during the study period, of which 135 cases with positive string test and PCR-amplified iucA (biosynthetic gene for the siderophore aerobactin) were defined as hvKP.10 String test was defined as positive when the bacterial colony can be extended to a viscous string at least 5 millimeters long from the surface of the agar plate.17 Secondly, in order to determine the prognostic factors related to the in-hospital mortality of the patients with hvKP infections, 135 patients with hvKP infections were divided into the survival group and the death group according to prognosis (Figure 1). Only the first strain isolated from the patients at the time of the first diagnosis of K. pneumoniae infection was collected, and subsequent isolates from the same patient were not incorporated into the study. Patient information including demographic characteristics, comorbidities, laboratory results, clinical manifestations, primary infection sites of hvKP, antimicrobial administration and clinical outcomes during hospitalization were obtained from medical records. Patients with incomplete information were excluded.
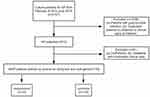 |
Figure 1 Flow chart for study inclusion. |
Typing of hvKP Strains and Their Capsule Type
PCR was used to detect the capsular serotypes of K1, K2, K5, K20, K54 and K57.18 MLST was tested by amplifying and sequencing the partial sequences of seven standard housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) and comparing them with KP MLST database to determine the allele types and STs of hvKP isolates (https://pubmlst.org/mlst).
Testing Virulence Genes of hvKP Strains
The presence of the virulence genes, including allantoin metabolism (allS), type 1 and type 3 fimbriae biosynthesis (fimH and mrkD), biosynthesis of lipopolysaccharide (ycfM and wabG), fucose synthesis (wcaG), mucoviscosity (prmpA, prmpA2 and magA), siderophore (entB, kfu, iroB, iroN, irp-1, irp-2, iutA, ybtS) and putative transporter (peg-344) was confirmed by PCR amplification and subsequent sequencing using the previously described methods and primers.6,19–21 Among them, several genes located on the pLVPK-like virulence plasmid, such as peg-344, PrmpA and PrmpA2 (regulators of the mucoid phenotype), have been proved to be related to the hypervirulence in the infection model in vivo.12,22 All the primer sequences for the mentioned genes above can be found in Additional file 1(Table S1).
Antimicrobial Susceptibility Testing and Antimicrobial Resistance Genes of hvKP Strains
The antimicrobial susceptibility test was performed initially by using AST-GN334 card on the VITEK 2 Compact (BioMérieux, Marcy-l’Étoile, France). The antimicrobial agents included piperacillin/tazobactam, ampicillin/sulbactam, ceftriaxone, ceftazidime, cefoperazone/sulbactam, cefepime, cefoxitin, ertapenem, imipenem, meropenem, tobramycin, amikacin, levofloxacin, aztreonam, sulfamethoxazole, minocycline, furazolidone and tigecycline. All the automatically-identified carbapenem resistant strains and tigecycline resistant strains were manually verified by standard broth microdilution methods on the basis of the 2018 Clinical and Laboratory Standards Institute (CLSI) guidelines.23 The results of tigecycline were judged according to the breakpoint of FDA Recognized Susceptibility Test Interpretive Criteria, with a minimum inhibitory concentration (MIC) ≥8.0 mg/L defined as resistant. The Escherichia coli ATCC 25922 strain was included as quality control. Genes encoding ESBLs (blaCTX-M-1 group, blaCTX-M-9 group, blaSHV and blaTEM) and carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48) were tested when the phenotypic test was positive, as previously reported.24 All positive PCR-amplified products were sequenced to validate their identity through BLAST website (https://blast.ncbi.nlm.nih.gov/blast.cgi).
Statistical Analysis
All statistical data were analyzed by SPSS statistical software (version 25, IBM Corporation, Armonk, NY, USA). The normal distribution of the statistical data was judged by One-Sample Kolmogorov–Smirnov test. The comparison of continuous variables was calculated by two simple t-test (normal distribution variables) or Mann–Whitney U-test (non-normal distribution variables). The comparison of the categorical variables was calculated by Chi-square test or Fisher’s exact test. The goodness fit of logistic regression model was evaluated by Hosmer-Lemeshow test. All variables with P-value <0.10 in the univariate analysis were included in the multivariate regression analysis to obtain the independent risk factors affecting the prognosis of patients. A 2-tailed P-value <0.05 was considered to be statistically significant. The odds ratio (OR) and its 95% confidence interval (CI) were calculated to evaluate the strength of any association.
Ethical Approval Statements
This retrospective study was approved by the Evaluation Committee and the Biomedical Ethics Committee of Chongqing Medical University (2020–460). In light of the retrospective and anonymous nature of the study, the Ethics Committee did not require written informed consent provided by participants.
Results
Clinical Characteristics of hvKP Infected Patients
In the present study, among the 613 cases of KP infections, a total of 135 patients were demonstrated to be infected by hvKP, with a prevalence rate of 22% (135/613). Sixteen of these patients died during hospitalization, with an in-hospital mortality rate of 11.9% (16/135). The detailed characteristics of the 135 patients diagnosed with hvKP are summarized in Table 1. The non-survivor group consisted of 4 females and 12 males, with an average age of 67.4 years. And the survivor group consisted of 44 females and 75 males, with an average age of 59.5 years. Nosocomial infections were predominant in both the two groups (p=0.188). The main underlying diseases in the non-survivor group included digestive system diseases (75%), respiratory diseases (56.3%) and hypertension (50%), while those in the survivor group included respiratory diseases (63.9%), digestive system diseases (62.2%) and Chronic kidney diseases (38.7%). However, there was no statistical difference in the underlying diseases between the survivor group and the non-survivor group. Respiratory tract infection, primary bacteremia and urinary tract infection were the main sources of infection in the two groups. The antimicrobial exposure within 4 weeks had a relatively small effect on mortality in both the surviving and non-surviving groups. The total length of hospital stays and post-culture of hospital stays were also similar in the two groups (p=0.878, 0.107, respectively). However, univariate analysis showed that the proportion of non-survivors admitted to ICU was much higher than that of the survivors (68.8% versus 34.5%, p=0.008).
 |
Table 1 Clinical Characteristics of Survivor and Non-Survivor Patients with hvKP |
Capsular Types and MLST of hvKP Strains
As shown in Table 2, K1 and K2 were the most prevalent capsular serotypes, accounted for 31.9% (43/135) and 25.2% (34/135). Moreover, there were 26.7% (36/135) of the strains that do not belong to the most common serotypes of K1, K2, K5, K20, K54, and K57. However, there was no statistical difference in the proportion of the above mentioned six serotypes between the non-survivor group and the survivor group.
 |
Table 2 Bacterial Characteristics of hvKP in Survivor and Non-Survivor Groups |
MLST analysis was implemented on all the 135 hvKP strains and 41 different STs were detected. Overall, ST23 was the most prevalent ST in the study (27.4%, 37/135), followed by ST86 (7.4%, 10/135), ST65 (5.2%, 7/135), ST412 (4.4%, 6/135), ST380 (3.7%, 5/135), ST29 (3.7%, 5/135). The above 6 STs accounted for 51.9% (70/135). Univariate analysis showed no statistical difference in the proportion of the above 6 STs between the dead and the survivors.
Virulence Genes of the hvKP Strains
Over 90% of the strains harbored entB, ycfM, peg-344, mrkD and fimH, accounting for 99.3%, 98.5%, 97.0%, 94.1% and 92.6%, respectively. Notably, the positive rate of iroN in hospitalized deaths was much higher than in survivors (87.5% versus 57.9%, p=0.023). In contrast, the positive rate of rmpA was higher in survivors than the dead (68.8% versus 89.9%, p=0.017). Whereas, our results did not find any significant difference in the frequency of other virulence genes between the two groups (Table 2).
Antimicrobial Susceptibility Testing of the hvKP Strains
Univariate analyses of the prognostic factors for mortality related to antimicrobial resistance are presented in Table 3. We found that the hvKP strains isolated from the non-survivor group exhibited higher rates of antimicrobial resistance for all the antibiotics (except tigecycline) than the hvKP strains isolated from the survivor group. Of note, the resistance rates for ampicillin/sulbactam, cefepime and aztreonam in the non-survivor group were significantly higher than those from the survivor group (37.5% versus 15.1%, p=0.028; 25% versus 5.9%, p=0.033; 25% versus 7.6%, p=0.049, respectively). Surprisingly, in the multiple logistic regression analysis, there was no statistical difference in the resistance rate of these β-lactam antibiotics between the two groups.
 |
Table 3 Antimicrobial Resistance of hvKP in Survivor and Non-Survivor Groups |
Antimicrobial Resistance Genes and MLST of ESBLs-Producing hvKP and CR-hvKP Strains
A total of 13 ESBLs-producing hvKP strains were isolated in this study. Among them, 12 strains carried blaCTX-M gene, 61.5% (8/13) of which were blaCTX-M-1 group and 30.8% (4/13) of which were blaCTX-M-9 group. The positive rates of blaSHV and blaTEM were 92.3% (12/13) and 23.1% (3/13), respectively. The 13 ESBLs-producing hvKP strains were divided into 10 different ST genotypes, of which 3 strains carrying both blaCTX-M-1 group and blaSHV belonged to ST23, and the others belonged to other different genotypes. The detailed information of 13 ESBLs-producing hvKP strains was in Additional file 1 (Table S2). Additionally, our study has identified five CR-hvKP strains harboring blaKPC-2 gene and several ESBLs genes (Table 4). All the five strains showed multiple resistances to the third- and fourth-generation cephalosporins, cephamycins and carbapenems, but were still susceptible to tigecycline. ST11 CR-hvKP were predominant, the remaining two strains belong to ST25 and ST2856, respectively. It is worth noting that one patient infected with ST25 KPC-producing CR-hvKP died of septic shock during hospitalization.
 |
Table 4 Clinical and Microbiological Characterization of CR-hvKP Isolates |
Prognostic Factors
Based on the study of the clinical characteristics of the patients and the microbiological characteristics of the strains, the variables in univariate analysis including age, admission to ICU, drug resistance to cefepime, aztreonam, ampicillin/sulbactam, and harboring iroN, rmpA gene were enrolled into the multivariate logistic regression analysis. The results displayed in Table 5 demonstrated that admission to ICU and harboring iroN gene were independent prognostic factors for in-hospital mortality of patients with hvKP infections (Hosmer Lemeshow test, P=0.799).
 |
Table 5 Prognostic Factors Associated with in-Hospital Mortality of Patients with hvKP Infections |
Discussion
Considering that hvKP tends to cause severe invasive diseases and thus elevate mortality, it is necessary to investigate the prevalence and mortality rate of hvKP and further reveal the predictive factors affecting prognosis. The prevalence and mortality rates of hvKP vary greatly depending on different regions. The prevalence rate of hvKP infection in our study is 22%, which is slightly lower than 24.5% to 47.5% previously reported in China, but much higher than 6% to 21% reported in other countries except China.5,25–27 Our consecutive study revealed that the in-hospital mortality rate for hvKP infection was 11.9%, slightly lower than the 17.5% reported in a previous retrospective study in Beijing, China, but far exceeds the in-hospital mortality of 2.3% reported in another study conducted in 10 cities of China.10,11
To our best knowledge, this is the first study to explore the prognostic factors for in-hospital mortality of patients with hvKP infections, which was defined by positive iucA and string test. Siderophores are beneficial for bacteria to obtain more free iron from their hosts.11,28 In addition to this traditional role, siderophores also play a crucial role in promoting the dissemination of bacteria, defensing against the host’s immune function of neutrophils and regulating the production of virulence genes.29–31 HvKP secrets four kinds of siderophores, which are enterobactin, salmochelin, yersiniabactin and aerobactin. Salmochelin was first identified as a C-glucosylated enterobactin produced by Salmonella enterica and uropathogenic Escherichia coli strains and other Enterobacteriaceae, including K. pneumoniae.32 Importantly, this C-glucosylated modification prevents salmochelin from being bound by lipocalin-2, which is capable of specifically sequestering the siderophore enterobactin with the highest affinity for iron, allowing salmochelin to suck more iron from its host.33 In a recent analysis of 2733 genomes of the K. pneumoniae complex, it was found that the trend of simultaneous existence of iro and iuc loci encoding aerobactin and salmochelin synthesis, respectively, was obvious, indicating the importance of coexistence of aerobactin and salmochelin to K. pneumoniae.34 The iroN gene encodes the outer-membrane receptor of salmochelin.32 Our statistics showed that in the presence of aerobactin, the presence of iroN may be an independent risk factor for hospital death in patients with hvKP infection. Magistro et al reported that iroN contributed to the formation of biofilm of extraintestinal pathogenic Escherichia coli.35 Moreover, the iroN was located on a large plasmid termed as pLVPK, which was demonstrated to be crucial to the pathogenicity of hvKP.36,37 Taken together, the above findings suggested that iroN is an independent prognostic factor for increased in-hospital mortality of patients with hvKP infections. However, another report described a fatal outbreak of ST11 CR-hvKP carrying pVir-CR-hvKP4 plasmid with an absence of prmpA and iroN genes.14 It may be partly caused by regional differences and different clone types of strains. In addition, more virulence genes that have not been evaluated in this study should be included to analyze their association with mortality in the future.
The increased in-hospital mortality associated with ICU admission could be explained by the following aspects. Firstly, the iroN gene showed a higher positive rate in patients with a history of ICU admission (OR =3.101, 95% CI =1.428–6.734; P=0.004). Secondly, patients admitted to ICU tend to exhibit multiple resistances to ampicillin/sulbactam, ceftazidime, cefepime, amikacin, aztreonam, which is bound to lead to inappropriate empirical treatment. In addition, more severe underlying illness and more fragile immune defenses in ICU patients, as well as the increase in invasive procedures, can be elements with fatal outcomes. Previous study has reported mortal outcomes in patients with an outbreak of hvKP infection admitted to the ICU.14 To sum up, patients admitted to ICU tend to have more devastating infection, coupled with inappropriate empirical treatment, so it is not surprising for the enhanced in-hospital mortality rate.
The resistance rate to β-lactam antibiotics in death group was slightly higher than that in the survival group in this study. However, the resistances to β-lactams did not exhibit a significant association with mortality, which is possibly due to the fitness cost, that is, the plasmids carrying antimicrobial resistance genes may be worse adaptable to hvKP.38 In addition, a total of 13 ESBLs-producing hvKP strains were isolated in our study, accounting for 9.6%, which is slightly lower than the 12.6–16.3% previously reported in China.11,26 It is troublesome that the dissemination of CTX-M and SHV gene through plasmids into hypervirulent variants from different STs. Because such strains could produce a variety of broad β-lactamases and ESBLs, which has a high level of resistance to multiple antimicrobial agents. Our findings reported five cases infected with KPC-producing CR-hvKP and one died of septic shock during hospitalization. All the 5 strains harbored blaKPC-2 and several other ESBLs genes. Previous studies have shown that blaKPC-2 gene can be transferred to hvKP by plasmid conjugation, which highly indicates that the horizontal transmission of blaKPC-2 in hvKP results in the generation of CR-hvKP in our study.39 As the dominant clone of CR-hvKP in our study, ST11’s ability to rapidly disseminate carbapenemases and its high pathogenicity were confirmed in a study that five patients successively died of severe pneumonia due to infection with ST11 KPC-producing hvKP in the ICU of a hospital in China.14 In addition, one patient infected with ST25 KPC-producing CR-hvKP died of septic shock during hospitalization in this study, which deserves more attention because ST25 CR-hvKP has also been spreading in a Chinese hospital before.40 In short, this must alert medical staffs to the possibility of horizontal transmission of resistant plasmids in hvKP, which may cause mortal outcomes in patients.
The generalizability of these results is subject to certain limitations. First, in light of hvKP being defined as positive for string test and iucA in this study and the relatively small sample size, the results of this study may not be applicable to other set of studies. Second, the results of this study were based on a retrospective analysis and data statistics, prospective studies and animal experiments were urgently needed to verify the prognostic factors of hvKP infection. Nevertheless, our findings have revealed the prevalence and in-hospital mortality of hvKP in southwestern China, and systematically analyzed the prognostic factors of death during hospitalization, which provides some predictive factors for clinicians to identify patients with poor prognosis in advance.
Conclusions
The prevalence and in-hospital mortality rate attributable to hvKP infection defined by positive string test and iucA were 22% and 11.9%, respectively. ICU admission and iroN carriage may be independent predictors of in-hospital mortality of hvKP-infected patients. Given the relatively high in-hospital mortality rate of patients with hvKP infections, early surveillance and better management are necessary for patients admitted to ICU and infected with iroN -harboring hvKP.
Abbreviations
ICU, intensive care unit; cKP, classic Klebsiella pneumoniae; hvKP, hypervirulent Klebsiella pneumoniae; CR-hvKP, carbapenem-resistant hypervirulent Klebsiella pneumoniae; pLVPK, pK2044-like plasmid; KPC, Klebsiella pneumoniae carbapenemase; OR, odds ratio; CI, confidence interval.
Acknowledgments
We thank the clinical laboratory of the Laboratory Department of the First Affiliated Hospital of Chongqing Medical University for providing experimental space and equipment.
Disclosure
The authors declare no conflicts of interest.
References
1. Paczosa M, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/mmbr.00078-15
2. Martin R, Bachman M. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi:10.3389/fcimb.2018.00004
3. Liu Y, Li B, Zhang Y, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014;58(9):5379–5385. doi:10.1128/AAC.02523-14
4. Fang C, Lai S, Yi W, Hsueh P, Liu K, Chang S. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. doi:10.1086/519262
5. Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect. 2016;22(2):154–160. doi:10.1016/j.cmi.2015.09.025
6. Liu C, Guo J. Characteristics of ventilator-associated pneumonia due to hypervirulent Klebsiella pneumoniae genotype in genetic background for the elderly in two tertiary hospitals in China. Antimicrob Resist Infect Control. 2018;7:95. doi:10.1186/s13756-018-0371-8
7. Alvarez D, Merino S, Tomas J, Benedi V, Alberti S, Moore RN. Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect Immun. 2000;68(2):953–955. doi:10.1128/iai.68.2.953-955.2000
8. Llobet E, Campos M, Gimenez P, Moranta D, Bengoechea J, Fang FC. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect Immun. 2011;79(9):3718–3732. doi:10.1128/iai.05226-11
9. Yeh K, Kurup A, Siu L, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. 2007;45(2):466–471. doi:10.1128/jcm.01150-06
10. Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. 2019;18(1):4. doi:10.1186/s12941-018-0302-9
11. Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. doi:10.1128/aac.01127-16
12. Russo T, Olson R, MacDonald U, Beanan J, Davidson B, Camilli A. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–3333. doi:10.1128/IAI.00430-15
13. Lai Y, Lu M, Hsueh P. Hypervirulence and carbapenem resistance: two distinct evolutionary directions that led high-risk clones to epidemic success. Expert Rev Mol Diagn. 2019;19(9):825–837. doi:10.1080/14737159.2019.1649145
14. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/s1473-3099(17)30489-9
15. Alba-Loureiro T, Munhoz C, Martins J, et al. Neutrophil function and metabolism in individuals with diabetes mellitus. Braz J Med Biol Res. 2007;40(8):1037–1044. doi:10.1590/s0100-879x2006005000143
16. Bruni E, Cazzetta V, Donadon M, et al. Chemotherapy accelerates immune-senescence and functional impairments of Vδ2(pos) T cells in elderly patients affected by liver metastatic colorectal cancer. J Immunother Cancer. 2019;7(1):347. doi:10.1186/s40425-019-0825-4
17. Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. doi:10.1093/cid/cit675
18. Turton J, Perry C, Elgohari S, Hampton C. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(5):541–547. doi:10.1099/jmm.0.015198-0
19. Kim D, Park B, Choi M, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. 2019;74(1):190–199. doi:10.1093/jac/dky397
20. Fu L, Huang M, Zhang X, et al. Frequency of virulence factors in high biofilm formation bla(KPC-2) producing Klebsiella pneumoniae strains from hospitals. Microb Pathog. 2018;116:168–172. doi:10.1016/j.micpath.2018.01.030
21. Russo T, Olson R, Fang C, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9). doi:10.1128/jcm.00776-18
22. Cheng H, Chen Y, Wu C, Chang H, Lai Y, Peng H. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192(12):3144–3158. doi:10.1128/jb.00031-10
23. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
24. Tian X, Sun S, Jia X, Zou H, Li S, Zhang L. Epidemiology of and risk factors for infection with extended-spectrum β-lactamase-producing carbapenem-resistant Enterobacteriaceae: results of a double case-control study. Infect Drug Resist. 2018;11:1339–1346. doi:10.2147/idr.S173456
25. Li J, Ren J, Wang W, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(4):679–689. doi:10.1007/s10096-017-3160-z
26. Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031–1041. doi:10.2147/idr.S161075
27. Namikawa H, Yamada K, Sakiyama A, et al. Clinical characteristics of bacteremia caused by hypermucoviscous Klebsiella pneumoniae at a tertiary hospital. Diagn Microbiol Infect Dis. 2019;95(1):84–88. doi:10.1016/j.diagmicrobio.2019.04.008
28. Kim D. Bacterial siderophores promote animal host iron acquisition and growth. Cell. 2018;175(2):311–312. doi:10.1016/j.cell.2018.09.020
29. Holden V, Breen P, Houle S, Dozois C, Bachman M. Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. mBio. 2016;7(5). doi:10.1128/mBio.01397-16
30. Lamont I, Beare P, Ochsner U, Vasil A, Vasil M. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2002;99(10):7072–7077. doi:10.1073/pnas.092016999
31. Saha P, Yeoh B, Olvera R, et al. Bacterial siderophores hijack neutrophil functions. J Immunol. 2017;198(11):4293–4303. doi:10.4049/jimmunol.1700261
32. Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003;100(7):3677–3682. doi:10.1073/pnas.0737682100
33. Skaar E. A precious metal heist. Cell Host Microbe. 2009;5(5):422–424. doi:10.1016/j.chom.2009.05.005
34. Lam M, Wyres K, Judd L, et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10(1):77. doi:10.1186/s13073-018-0587-5
35. Magistro G, Hoffmann C, Schubert S. The salmochelin receptor IroN itself, but not salmochelin-mediated iron uptake promotes biofilm formation in extraintestinal pathogenic Escherichia coli (ExPEC). Int J Med Microbiol. 2015;305:435–445. doi:10.1016/j.ijmm.2015.03.008
36. Chen Y, Chang H, Lai Y, Pan C, Tsai S, Peng H. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi:10.1016/j.gene.2004.05.008
37. Struve C, Roe C, Stegger M, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio. 2015;6(4):e00630. doi:10.1128/mBio.00630-15
38. Long D, Zhu LL, Du FL, et al. Phenotypical profile and global transcriptomic profile of Hypervirulent Klebsiella pneumoniae due to carbapenemase-encoding plasmid acquisition. BMC Genomics. 2019;20(1):480. doi:10.1186/s12864-019-5705-2
39. Siu L, Huang D, Chiang T. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis. 2014;14:176. doi:10.1186/1471-2334-14-176
40. Li J, Huang Z, Yu T, et al. Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. 2019;19(1):219. doi:10.1186/s12866-019-1593-5
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
