Back to Journals » International Journal of General Medicine » Volume 11
Clinical and microbiological characteristics and occurrence of Klebsiella pneumoniae infection in Japan
Authors Ikeda M, Mizoguhi M, Oshida Y, Tatsuno K, Saito R , Okazaki M, Okugawa S , Moriya K
Received 1 March 2018
Accepted for publication 9 May 2018
Published 13 July 2018 Volume 2018:11 Pages 293—299
DOI https://doi.org/10.2147/IJGM.S166940
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Mahoko Ikeda,1,2,* Miyuki Mizoguchi,1,* Yukie Oshida,3 Keita Tatsuno,1 Ryoichi Saito,4 Mitsuhiro Okazaki,5 Shu Okugawa,2 Kyoji Moriya1,2
1Department of Infection Control and Prevention, The University of Tokyo Hospital, Hongo, Bunkyo-ku, Tokyo, Japan; 2Department of Infectious Diseases, The University of Tokyo Hospital, Hongo, Bunkyo-ku, Tokyo, Japan; 3Division of Infectious Diseases, Shizuoka Cancer Center, Simonagakubo, Nagaizumi-chou, Suntou-gun, Shizuoka, Japan; 4Department of Microbiology and Immunity, Graduate School of Health Care Sciences, Tokyo Medical and Dental University, Yushima, Bunkyo-ku, Tokyo, Japan; 5Department of Medical Technology, School of Health Sciences, Tokyo University of Technology, Kamata, Ota-ku, Tokyo, Japan
*These authors contributed equally to this work
Purpose: Klebsiella pneumoniae is a pathogen that causes pneumonia and urinary tract infection. Hypervirulent K. pneumoniae strains often show hypermucoviscosity, are of the K1 or K2 serotype, and harbor the rmpA and magA genes. However, the differences in the prevalence of K. pneumoniae with these hypervirulent characteristics between the infection and colonization status are not well understood. Therefore, in this study, we compared the clinical and microbiological characteristics of K. pneumoniae isolated from urine or sputum samples of cases of infection and colonization.
Patients and methods: This retrospective study was conducted at a tertiary care teaching hospital in Tokyo, Japan. Patients whose sputum or urine tested positive for the presence of K. pneumoniae isolates were randomly included in the study. Clinical and microbiological data were collected from medical records.
Results: Of the 130 cases investigated, 68 and 62 cases showed the presence of K. pneumoniae in the sputum and urine, respectively. There were 49 infection cases, including 21 in the sputum group and 28 in the urine group. The infections were not accompanied by liver abscess. Of the 130 K. pneumoniae isolates, 25 (19.2%) showed capsular serotype K1 or K2, whereas 33 (25.4%) showed hypermucoviscosity. The prevalence of virulence genes magA, allS, rmpA, mrkD, uge, kfu-BC, and wabG was 10% (all in K1), 13.1%, 16.9%, 85.4%, 79.2%, 36.9%, and 91.5%, respectively. In both the sputum and urine groups, there was no difference in the characteristics of patients with infection and those with colonization. Analysis of microbiological characteristics revealed that only rmpA was significantly more frequent in the infection cases than in the colonization/asymptomatic cases in both the sputum and urine groups.
Conclusion: The rmpA-positive K. pneumoniae isolates were dominant in the infection cases compared with those in the colonization/asymptomatic cases, suggesting that rmpA may play a crucial role in the development of urinary tract infection and pneumonia.
Keywords: Klebsiella pneumoniae, hypermucoviscosity, rmpA, capsular polysaccharide serotype, infection, colonization, pneumonia, urinary tract infection
Introduction
Klebsiella pneumoniae is the infectious agent in various diseases, such as bacteremia, meningitis, pneumonia, and urinary tract infection (UTI). A hypervirulent variant of K. pneumoniae that causes severe liver abscesses and bacteremia emerged in the 1980s,1–3 which shows the main characteristics of hypermucoviscosity and tendency to cause severe community-acquired infection.4 This variant was initially mainly reported in Asian countries such as Taiwan5 and South Korea,6 but has now spread to countries outside Asia.7–9 This new variant often harbors the K1 or K2 capsular polysaccharide.4
In addition to the K1 or K2 capsular polysaccharide and the hypermucoviscous phenotype, several genes have been reported as severity determinants, including magA and rmpA, based on studies comparing cases with severe infection to those with localized infection, and in murine lethality experiments.10,11 The magA gene, which encodes a polymerase involved in capsule synthesis, first emerged as one of the candidate virulence genes,12 but is now recognized as a K1 surrogate marker.13,14 Moreover, rmpA, which regulates capsular polysaccharide synthesis, was shown to be responsible for virulence in clinical studies10,13,15 and in murine models.16,17 The kfu gene, which mediates the uptake of ferric iron, and allS, encoding the activator of the allantoin regulon,18 were also considered as virulence factors in a murine intraperitoneal challenge model.19,20 Other reported virulence factors of K. pneumoniae include the wabG gene, which plays a role in formation of the outer core lipopolysaccharide, and the uge gene, which encodes a uridine diphosphate galacturonate 4-epimerase and is required for biosynthesis of the capsule and smooth lipopolysaccharide.21 However, the prevalence of these genes did not significantly differ between UTI cases and cases without infection.22 The mrkD gene, which encodes type 3 fimbrial adhesion protein, was reported as a factor related to biofilm formation,23 but its influence on virulence in a clinical setting remains unclear.24
Thus, the factors that cause severe infections, such as bacteremia and liver abscess, have been widely investigated both in clinical studies and in animal models. Moreover, some epidemiological studies of hypervirulent K. pneumoniae strains causing other types of infections, such as pneumonia and UTI, have also been reported.22,25 Furthermore, some of the virulence factors of K. pneumoniae pneumonia associated with bacteremia in Japan were identified.26 Despite this recent focus on the virulence factors underlying severe bacterial infection and their relationship with infection severity, the difference in the prevalence of the hypervirulent K. pneumoniae strains and other strains and the characteristics between infection and colonization/asymptomatic status are still poorly understood.
Therefore, we performed a retrospective study to investigate the prevalence of the hypermucoviscous phenotype and virulent K. pneumoniae strains in a tertiary care teaching hospital in Japan and to determine the differences in infection status with respect to patient background and microbiological characteristics. This study can contribute to gaining a deeper understanding of the epidemiology of virulent K. pneumoniae strains and the virulence factors involved in the development of K. pneumoniae infection.
Patients and methods
Patients and setting
This retrospective study was conducted at the University of Tokyo Hospital, a 1,217-bed tertiary care teaching hospital in Tokyo, Japan. We randomly selected 130 patients among those who tested positive for the presence of K. pneumoniae isolates in their sputum or urine from January 2011 to November 2012. One isolate from the sputum or urine was included from each patient. Patient data, clinical symptoms, and microbiological data were retrieved from the medical records.
Data collection and definitions
Collected patient data included age, sex, underlying disease (diabetes mellitus, malignancy, neutropenia, collagen disease, cirrhosis, chronic kidney disease, and chronic pulmonary disease), and use of immunosuppressant. The presence of intravascular catheters, including peripheral vessel catheters, central venous catheters, and arterial vessel catheters, as well as urinary catheters was reviewed. History of residence in a nursing home and antibiotic use within a month were also reviewed. The samples obtained within 48 hours after admission were defined as community isolates; the other samples were defined as hospital isolates.
The cases were divided into infection and colonization cases. Infection was defined depending on the type of sample that showed the presence of K. pneumoniae; ie, pneumonia in cases from whom sputum was obtained and UTI in cases from whom urine was obtained. Definitions of these infections were compliant with the relevant guidelines.27,28 Pneumonia due to K. pneumoniae was defined as 1) manifestation of clinical symptoms such as fever (>37.5°C) and purulent sputum (assessed by Gram staining or appearance of sputum), 2) compatible radiological findings, and 3) dominant detection of K. pneumoniae in sputum culture. Pneumonia cases for which the samples did not meet all of these criteria were defined as colonization cases. UTI due to K. pneumoniae was defined as 1) manifestation of clinical symptoms such as fever (>37.5°C) and pain during urination, 2) positive leukocyte esterase test in urinary analysis, and 3) single or dominant detection of K. pneumoniae in urine culture. UTI cases for which the samples did not meet all of these criteria were defined as asymptomatic cases.
Microbiological procedures
All isolates were identified using the WalkAway system (Siemens, Berlin, Germany). Antimicrobial susceptibility was determined with the WalkAway system and the standard criteria of the Clinical and Laboratory Standards Institute guide M100-S27.29 The hypermucoviscosity of K. pneumoniae was assessed with the string test, in which a viscous string >5-mm long was considered positive.24,30 The capsular polysaccharide serotype (K1 and K2) was determined by polymerase chain reaction (PCR) for serotype-specific targets within the K1 and K2 cps clusters, as described previously.31 To detect virulence-associated genes, such as magA, allS, rmpA, mrkD, uge, kfu-BC, and wabG, the genomic DNA of K. pneumoniae was extracted and PCR was performed with the specific primers, according to a previous report.32
Statistical analyses
Categorical data were analyzed using the two-tailed Fisher’s exact test and nonparametric data were analyzed using the Mann–Whitney U-test. Differences with p<0.05 were considered significant. All statistical analyses were performed using JMP Pro version 11 software (SAS Institute, Cary, NC, USA).
Ethical considerations
This study was approved by the research ethics committee of the University of Tokyo Hospital. The requirement to obtain written informed consent from each patient was waived because this was an observational retrospective study. The data were analyzed anonymously.
Results
Clinical characteristics of patients
During the study period, 12,467 urine and 4,773 sputum samples were submitted to the clinical laboratory. Among these, 4,093 urine and 1,989 sputum samples were positive for bacteria or fungi. K. pneumoniae was detected in the sputum of 322 cases and in the urine of 288 cases. We randomly enrolled 130 cases (68 in the sputum group and 62 in the urine group) and divided them into infection cases and colonization/asymptomatic cases.
The sputum group comprised 21 pneumonia (infection) and 47 colonization cases. The patients’ characteristics in the infection and colonization groups were similar (Table 1). Two infection cases were complicated by bacteremia. No liver abscess accompanied pneumonia.
  | Table 1 Characteristics of patients in whom Klebsiella pneumoniae was isolated from the sputum |
The urine group comprised 28 UTI (infection) and 34 asymptomatic cases, and the patients’ characteristics were similar between the two groups (Table 2). Bacteremia was present in four cases. No liver abscess was detected in any of the UTI cases.
  | Table 2 Characteristics of patients in whom Klebsiella pneumoniae was isolated from the urine |
Virulence genes and antimicrobial susceptibility of K. pneumoniae isolates
Of the 130 K. pneumoniae isolates, 25 (19.2%) exhibited capsular serotype K1 or K2 and 33 (25.4%) showed hypermucoviscosity. The prevalence of virulence-associated genes in the sputum and urine groups is shown in Tables 3 and 4. In both the sputum and urine groups, the prevalence of rmpA in the infection cases was significantly higher than that in the colonization/asymptomatic cases (p=0.016 in the sputum group and p=0.037 in the urine group). Prevalence of the other genes did not differ significantly between the infection and colonization/asymptomatic cases in both the sputum and urine groups.
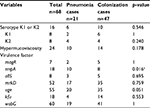  | Table 3 Bacterial characteristics of Klebsiella pneumoniae isolated from pneumonia cases Note: aStatistically significant. |
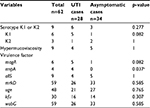  | Table 4 Bacterial characteristics of Klebsiella pneumoniae isolated from urinary tract infection cases Note: aStatistically significant. Abbreviation: UTI, urinary tract infection. |
Of the 130 isolates, antimicrobial susceptibility data for one strain were missing. Except for this isolate, all of the isolates showed 100% sensitivity to ceftazidime, cefozopran, meropenem, gentamicin, and levofloxacin. No extended-spectrum beta-lactamase (ESBL)-producing strains were detected.
Discussion
K. pneumoniae is the etiological agent of pneumonia and UTI. Although the pathogenesis and virulence factors of K. pneumoniae have been widely studied, the differences between infectious cases and colonization/asymptomatic cases are not well understood. Therefore, in this study, we compared the clinical and microbiological characteristics between infection cases and colonization cases in a tertiary care teaching hospital in Japan. We determined the prevalence of hypermucoviscosity, specific serotypes, and virulence genes in K. pneumoniae isolated from the sputum and urine obtained from selected patients. We also showed the significant dominance of rmpA in infection cases in both the sputum and urine groups.
Diabetes mellitus has been reported as a risk factor for hypervirulent K. pneumoniae infection.4,33 However, we did not find a difference in the prevalence of diabetes mellitus between the infection and colonization/asymptomatic cases.
The infection and colonization/asymptomatic groups showed a similar prevalence of serotypes and the hypermucoviscous phenotype; 25.4% and 19.2% of all isolates had the hypermucoviscous phenotype and the K1/K2 serotypes, respectively. Although the epidemiology of hypermucoviscous K. pneumoniae in various types of infection in Japan is not clear, ~10% of bacteremic K. pneumoniae were reported to exhibit the hypermucoviscous phenotype34 and 18.3% of K. pneumoniae isolates causing pneumonia belonged to the K1 and K2 serotypes.26 In Taiwan, approximately half of the K. pneumoniae isolates were found to belong to serotype K1/K2 in patients with community-acquired bacterial pneumonia25,35 and in nasopharynx carriers.35 In China, 33% of the isolates from hospitalized patients were reported to be hypermucoviscous.36 Outside Asia, infection with serotype K1/K2 or hypermucoviscous K. pneumoniae appears to be quite rare. In Europe, 1.1% of 1090 isolates belonged to the K1-CC23 serotype in the UK,37 and 5.4% of 878 isolates were hypermucoviscous, whereas 0.3% were rmpA-positive in Spain.38 In the USA, 6.3% of the 64 isolates obtained from blood cultures were found to be rmpA- or magA-positive.39 These reports indicate that the prevalence of specific serotypes and the hypermucoviscous phenotype vary geographically.
Of the virulence genes investigated in the present study, rmpA was significantly dominant in the infection cases. Indeed, the K1/K2 strains, previously known as virulent strains, were shown to be avirulent without rmpA or the hypermucoviscous phenotype in a murine model.40 Among the non-K1/K2 strains, rmpA-positive strains showed high virulence in murine lethality tests.41 Moreover, in Taiwan, 85.7% and 87.0% of non-K1/K2 isolates were found to be rmpA-positive in cases of liver abscess and bacteremia, respectively.25,40 Although some rmpA-positive isolates did not exhibit the hypermucoviscous phenotype, these isolates were more virulent than the rmpA-negative and nonhypermucoviscous isolates.40 Lin et al22 reported that the prevalence of rmpA was significantly higher in the urinary isolates of UTI cases than that in the fecal isolates obtained from healthy people in Taiwan.
The rmpA gene regulates capsule polysaccharide biosynthesis.16 Three types of rmpA genes have been reported to date: chromosomally located rmpA, rmpA on a plasmid, and rmpA2 on a plasmid.24 Although the primers used in this study could only detect plasmid-mediated rmpA and rmpA2, the strains possessing chromosomally located rmpA appear to be much less prevalent than those possessing plasmid-mediated rmpA/A2.24 Hypercapsule synthesis regulated by rmpA contributes to preventing the phagocytosis and opsonophagocytosis of K. pneumoniae by the host immune cells and inhibits complement-mediated lysis and opsonization.42,43 Thus, rmpA regulation is associated with the escape of K. pneumoniae from immune responses,44 indicating a potentially crucial role in the development of K. pneumoniae infection. Although the prevalence of other reported virulence genes, such as magA, allS, mrkD, uge, kfu, and wabG, did not differ significantly between the infection and colonization groups, we cannot exclude the possibility that these genes might also have some effect in the development of K. pneumoniae infection, especially since not all of the infection cases were associated with an rmpA-positive strain.
No antimicrobial multiresistant strain was detected in our study. A previous study24 reported that hypermucoviscous strains are generally susceptible to antimicrobials except for ampicillin; however, ESBL-producing strains and carbapenem-resistant strains are emerging, especially in China.45,46 Although the prevalence rate of ESBL-producing K. pneumoniae is still low in Japan,47 multi-antimicrobial resistance among hypermucoviscous K. pneumoniae strains may become a future threat.
There are several limitations of this study. First, because this was a retrospective study, it is unknown whether the rmpA-positive isolates in the colonization/asymptomatic group eventually caused infections. Second, this was a single-center analysis and the sputum and urine samples were randomly selected; therefore, not all the samples collected during this time period were analyzed. Thus, a multicenter study is warranted, especially considering that the prevalence of the hypervirulent variant differs from region to region.
Conclusion
In conclusion, the prevalence of hypermucoviscous K. pneumoniae was found to be 25.4%, and rmpA was detected at significantly higher rates in the infection cases than in the colonization/asymptomatic cases in both the sputum and urine groups. The prevalence of serotypes and the hypermucoviscous phenotype is highly variable across studies according to the geographical region. Nevertheless, our study suggests that the rmpA gene may play a crucial role in the development of UTI and pneumonia.
Acknowledgments
We thank all of the microbiological technologists at the University of Tokyo Hospital for their technical support. This work was presented at the Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) 2015, September 17–21, 2015, San Diego, CA, USA.
Disclosure
The authors report no conflicts of interest in this work.
References
Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146(10):1913–1916. | ||
Hu BS, Lau YJ, Shi ZY, Lin YH. Necrotizing fasciitis associated with Klebsiella pneumoniae liver abscess. Clin Infect Dis. 1999;29(5):1360–1361. | ||
Chou FF, Kou HK. Endogenous endophthalmitis associated with pyogenic hepatic abscess. J Am Coll Surg. 1996;182(1):33–36. | ||
Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881–887. | ||
Lee SS, Chen YS, Tsai HC, et al. Predictors of septic metastatic infection and mortality among patients with Klebsiella pneumoniae liver abscess. Clin Infect Dis. 2008;47(5):642–650. | ||
Kim JK, Chung DR, Wie SH, Yoo JH, Park SW; Korean Study Group for Liver Abscess. Risk factor analysis of invasive liver abscess caused by the K1 serotype Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2009;28(1):109–111. | ||
Nadasy KA, Domiati-Saad R, Tribble MA. Invasive Klebsiella pneumoniae syndrome in North America. Clin Infect Dis. 2007;45(3):e25–e28. | ||
Decré D, Verdet C, Emirian A, et al. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol. 2011;49(8):3012–3014. | ||
Turton JF, Englender H, Gabriel SN, Turton SE, Kaufmann ME, Pitt TL. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol. 2007;56(Pt 5):593–597. | ||
Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358. | ||
Yeh KM, Kurup A, Siu LK, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. 2007;45(2):466–471. | ||
Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. | ||
Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2006;193(5):645–654. | ||
Yeh KM, Chang FY, Fung CP, Lin JC, Siu LK. magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of Klebsiella pneumoniae serotype K1. J Med Microbiol. 2006;55(Pt 6):803–804. | ||
Yu VL, Hansen DS, Ko WC, et al; International Klebseilla Study Group. Virulence characteristics of Klebsiella and clinical manifestations of Klebsiella pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13(7):986–993. | ||
Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192(12):3144–3158. | ||
Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157(Pt 12):3446–3457. | ||
Chou HC, Lee CZ, Ma LC, Fang CT, Chang SC, Wang JT. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun. 2004;72(7):3783–3792. | ||
Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2008;197(12):1717–1727. | ||
Ma LC, Fang CT, Lee CZ, Shun CT, Wang JT. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J Infect Dis. 2005;192(1):117–128. | ||
Regué M, Hita B, Piqué N, et al. A gene, uge, is essential for Klebsiella pneumoniae virulence. Infect Immun. 2004;72(1):54–61. | ||
Lin WH, Wang MC, Tseng CC, et al. Clinical and microbiological characteristics of Klebsiella pneumoniae isolates causing community-acquired urinary tract infections. Infection. 2010;38(6):459–464. | ||
Jagnow J, Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149(Pt 9):2397–2405. | ||
Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. | ||
Lin YT, Jeng YY, Chen TL, Fung CP. Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: clinical and microbiological characteristics in Taiwan, 2001–2008. BMC Infect Dis. 2010;10:307. | ||
Ito R, Shindo Y, Kobayashi D, et al. Molecular epidemiological characteristics of Klebsiella pneumoniae associated with bacteremia among patients with pneumonia. J Clin Microbiol. 2015;53(3):879–886. | ||
Mandell LA, Wunderink RG, Anzueto A, et al; Infectious Diseases Society of America; American Thoracic Society. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72. | ||
Hooton TM, Bradley SF, Cardenas DD, et al; Infectious Diseases Society of America. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin Infect Dis. 2010;50(5):625–663. | ||
Standards. CaL. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-first Informational Supplement M100-S21. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2011. | ||
Kawai T. Hypermucoviscosity: an extremely sticky phenotype of Klebsiella pneumoniae associated with emerging destructive tissue abscess syndrome. Clin Infect Dis. 2006;42(10):1359–1361. | ||
Vila A, Cassata A, Pagella H, et al. Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol J. 2011;5:107–113. | ||
Brisse S, Fevre C, Passet V, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. 2009;4(3):e4982. | ||
Cheng DL, Liu YC, Yen MY, Liu CY, Wang RS. Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch Intern Med. 1991;151(8):1557–1559. | ||
Togawa A, Toh H, Onozawa K, et al. Influence of the bacterial phenotypes on the clinical manifestations in Klebsiella pneumoniae bacteremia patients: a retrospective cohort study. J Infect Chemother. 2015;21(7):531–537. | ||
Lin YT, Wang YP, Wang FD, Fung CP. Community-onset Klebsiella pneumoniae pneumonia in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front Microbiol. 2015;9:122. | ||
Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. | ||
Turton JF, Payne Z, Coward A, et al. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and “non-hypervirulent” types ST147, ST15 and ST383. J Med Microbiol. 2018;67(1):118–128. | ||
Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect. 2016;22(2):154–160. | ||
Chou A, Nuila RE, Franco LM, Stager CE, Atmar RL, Zechiedrich L. Prevalence of hypervirulent Klebsiella pneumoniae-associated genes rmpA and magA in two tertiary hospitals in Houston, TX, USA. J Med Microbiol. 2016;65(9):1047–1048. | ||
Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62(1):1–6. | ||
Lin HA, Huang YL, Yeh KM, Siu LK, Lin JC, Chang FY. Regulator of the mucoid phenotype A gene increases the virulent ability of extended-spectrum beta-lactamase-producing serotype non-K1/K2 Klebsiella pneumonia. J Microbiol Immunol Infect. 2016;49(4):494–501. | ||
Lin JC, Chang FY, Fung CP, et al. High prevalence of phagocytic-resistant capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect. 2004;6(13):1191–1198. | ||
Domenico P, Salo RJ, Cross AS, Cunha BA. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect Immun. 1994;62(10):4495–4499. | ||
Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. | ||
Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. | ||
Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2015;60(1):709–711. | ||
Shibasaki M, Komatsu M, Sueyoshi N, et al. Community spread of extended-spectrum beta-lactamase-producing bacteria detected in social insurance hospitals throughout Japan. J Infect Chemother. 2016;22(6):395–399. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
