Back to Journals » Cancer Management and Research » Volume 10
Clinical and biological implications of IDH1/2 in acute myeloid leukemia with DNMT3Amut
Authors Zhang XP, Shi JL, Zhang JL , Yang XR, Zhang GQ, Yang SY, Wang J, Ke XY, Fu L
Received 20 November 2017
Accepted for publication 27 April 2018
Published 6 August 2018 Volume 2018:10 Pages 2457—2466
DOI https://doi.org/10.2147/CMAR.S157632
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Xinpei Zhang,1 Jinlong Shi,2 Jilei Zhang,1 Xinrui Yang,1 Gaoqi Zhang,1 Siyuan Yang,1 Jing Wang,1 Xiaoyan Ke,1 Lin Fu1
1Department of Hematology and Lymphoma Research Center, Peking University, Third Hospital, Beijing 100191, China; 2Department of Biomedical Engineering, Chinese PLA General Hospital, Beijing 100853, China
Purpose: The incidence of DNMT3A mutations in acute myeloid leukemia (AML) is quite high and often confers a poorer prognosis. Another common gene involved in AML is IDH1/2. However, the influence of IDH1/2 mutations on outcomes in DNMT3A-mutated patients remains unknown. This study aims to determine the effect of IDH1/2mut on the prognosis in patients with DNMT3A-mutated AML.
Patients and methods: We screened patients from The Cancer Genome Atlas database and selected 51 patients with AML and the DNMT3A mutation, among which 16 patients (31.4%) had both DNMT3A and IDH1/2mut.
Results: Among our sample, 11 cases had the IDH1 mutation (21.7%), and 5 cases had the IDH2 mutation (9.8%). Patients in the DNMT3AmutIDH1/2wild group showed a greater number of NPM1 mutation (P=0.022), and higher event-free survival (EFS) and overall survival (OS) after hematopoietic stem cell transplantation (HSCT) (P=0.010 and P=0.007, respectively). Patients in the DNMT3AmutIDH1/2mut group showed no increase in EFS or OS after HSCT or chemotherapy. Other factors, like white blood cells, bone marrow blasts, peripheral blood blasts, and mutated recurrent gene numbers had no significant influence on EFS and OS.
Conclusion: The IDH1/2 gene had little influence on the prognosis of patients with the DNMT3A mutation. For patients in the DNMT3AmutIDH1/2wild group, HSCT had a more favorable therapeutic effect. For patients with DNMT3A and IDH1/2mut, chemotherapy and HSCT appeared to have similar efficacy.
Keywords: acute myeloid leukemia, molecular mutations, prognosis, next-generation sequencing
Introduction
Acute myeloid leukemia (AML) is an aggressive hematological disease that is characterized by the overproduction of early myeloid precursor cells, often to the exclusion of other cell lines. It leads to anemia, thrombocytopenia, and neutropenia,1,2 all of which typically have poor outcomes. Recently, some significant improvements have been achieved in our understanding of AML biology and genetics. These fundamental discoveries are now being translated into new diagnostic and therapeutic strategies for this disease.3
One of the commonly mutated genes in AML is DNMT3A. It encodes a DNA methyltransferase that is localized in the cytoplasm and nucleus, and plays a role in de novo methylation. The prevalence of mutations in DNMT3A ranges from 18% to 36% in AML.4,5 Hajkova et al found significantly lower levels of global DNA methylation in AML patients with DNMT3A mutations, and this hypomethylation correlated with higher relapse rates and poorer overall survival (OS).6 Many recent studies have also suggested that the DNMT3A mutation should be considered a poor prognostic factor in AML.5–9 The frequencies of IDH1 and IDH2 mutations in AML are approximately 6–16% and 8–19%, respectively.10,11 IDH is an essential metabolic enzyme that catalyzes the oxidative decarboxylation of isocitrate to α-KG. The IDH1/2 gene mutations give rise to reduced levels of α-KG and increased levels of 2HG.10,12 In AML, increasing cellular 2HG levels will inhibit α-KG-dependent enzymes that are important for the demethylation of DNA.10,13,14 Studies have shown that IDH1/2mut display global DNA hypermethylation, and in hematopoietic stem cells, expression of mutant IDH1/2 increases the expression of stem cell markers and impaired myeloid differentiation.15 A recent meta-analysis indicated that IDH1 mutations confer a poorer OS and event-free survival (EFS). However, for IDH2, some differences were noted. The IDH2R140 mutations were associated with better OS among younger cases, whereas outcome was poor for patients with the IDH2R172 mutation.10,16
Mutations in DNMT3A and IDH1/2 have a significant effect on the prognosis of AML patients. They are both involved in the epigenetic regulation of transcription, particularly, alterations in DNA methylation. In addition, these genes both show a high co-mutation rate. However, whether the pattern of association between DNMT3Amut and IDH1/2mut suggests an interplay in the prognosis of AML remains unknown. This study aims to determine whether IDH1/2 influences the outcomes of AML patients with the DNMT3A mutation, and its effects.
Patients and methods
Patients
Fifty-one patients diagnosed with AML and the DNMT3A mutation were enrolled in the study. Experimental data were derived from The Cancer Genome Atlas database (https://cancergenome.nih.gov/). Data on demographic and molecular characteristics of patients with AML, EFS, OS, etc. were also collected and analyzed. Recurrent genetic mutations were detected by next-generation sequencing. Patients were treated in accordance with national comprehensive cancer network guidelines (https://www.nccn.org), with an emphasis on enrollment in therapeutic clinical trials wherever possible. Patients with poor risk received allogeneic hematopoietic stem cell transplantation (allo-HSCT) if they were medically fit for the associated risks of transplantation, and if a suitably matched donor was available. Many patients with intermediate risk also underwent allo-HSCT at some point during the course of the disease.17 Four patients with intermediate risk received autogenic hematopoietic stem cell transplantation (auto-HSCT). Nineteen patients received allo-HSCT, among which 9 received sibling allo-HSCT (2 high-risk patients and 7 intermediate-risk patients), 6 received matched unrelated donor (MUD) HSCT (1 high-risk patient and 5 intermediate-risk patients), 3 received auto- and MUD HSCT (1 high-risk patient and 2 intermediate-risk patients), and 1 intermediate-risk patient received auto- and sibling allo-HSCT.
To further support the analysis, we used the data of patients in the Clinseq cohort.18 There were 63 patients with the DNMT3A mutation in the Clinseq cohort who were diagnosed between February 1997 and August 2014. Bone marrow or peripheral blood samples were obtained at the time of diagnosis. These patients were treated with intensive induction regimens, including anthracyclines and cytosine arabinoside, according to national guidelines.19
Statistical analysis
The primary study endpoints were EFS and OS. The EFS was defined as that time from the date of diagnosis to removal from the study, owing to the absence of complete remission, relapse, or death. The OS was defined as that time from the date of diagnosis to death by any cause. For patients in the Clinseq cohort, we considered OS as the primary outcome.
The demographics and characteristics of the subjects were summarized using descriptive statistics. Intergroup difference was performed using the Student’s t-test and chi-square test. Survival analyses were performed using the Kaplan–Meier method. A two-sided P-value <0.05 was considered statistically significant. All statistical analyses were performed with the SPSS software 20.0 (IBM Corporation, Armonk, NY, USA).
Results
Demographic and biological characteristics
The mutation rates of IDH are shown in Figure 1. Sixteen patients had either the IDH1 or IDH2 mutation, accounting for 31.4%. The IDH1 mutations were observed more frequently than IDH2 mutations. Eleven patients with the IDH1 mutation had a mutation at R132. Three patients with the IDH2-mutation had an IDH2R140 mutation, and the other 2 had an IDH2R172 mutation.
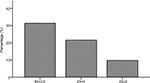  | Figure 1 Mutation rate of IDH. Note: Total mutation frequency of IDH1/2 was 31.4% (16 cases); 21.6% (11 cases) for IDH1 and 9.8% (5 cases) for IDH2. |
The demographic and biological characteristics and the intergroup differences are summarized in Table 1. The median age among all 51 patients was 58 years (range: 21–81 years); 27 patients (52.9%) were younger than 60 years. The subjects included 24 males (47.1%) and 27 females (52.9%). In AML FAB, patients with a subtype of M1, 9 (17.6%) of them had DNMT3A and IDH1/2 double mutations while only 4 (7.8%) had only DNMT3Amut (P=0.001). The median white blood cell (WBC) count was 45×109/L (range: 1.2×109/L–298.4×109/L), and 19 cases (37.3%) had counts ≥ 50×109/L WBC. The median percentages of bone marrow (BM) blasts and peripheral blood (PB) blasts were 76% (range: 32–100%) and 36% (range, 0–97%), respectively. The percentages of BM blasts and PB blasts in 30 cases (58.8%) and 15 cases (30.0%), respectively, were ≥ 70%. Twenty-eight patients (54.9%) relapsed after receiving treatment. However, these characteristics showed no significant differences between the DNMT3AmutIDH1/2wild and DNMT3AmutIDH1/2mut groups.
The mutation status of the genes under investigation is summarized in Table 1. All genes were associated with AML and had a mutation rate > 5% in our cohort. The most frequently mutated gene was NPM1, with 28 mutated cases (54.9%), most of which were concentrated in the DNMT3AmutIDH1/2wild group (P=0.022). Other mutations, including those of FLT3-ITD/TKD, TET2, SMC3, NRAS, KRAS, CEBPA, PTPN11, U2AF1, SMC1A, and RAD21, showed no significant intergroup differences (P> 0.05). Among the 51 cases, 23 patients (45.1%) underwent HSCT. In the DNMT3AmutIDH1/2mut group, 11 cases accepted HSCT, whereas 5 did not. The proportion of HSCT acceptance was significantly higher in patients with double mutations (P=0.022).
Comparison of EFS and OS between the DNMT3AmutIDH1/2wild and DNMT3AmutIDH1/2mut groups
In order to determine which patients were more likely to be affected by the IDH1/2 mutation, we stratified the data by several aspects, such as age, WBC count, percentages of BM blasts and PB blasts, mutated recurrent gene numbers, and frequently mutated genes, such as FLT3 and NPM1. The results of the Kaplan–Meier analysis for EFS and OS are summarized in Table 2. Although the median survival time seemed longer in the double mutation group in several aspects, such as age <60 years, BM blasts or PB blasts <70%, FLT3 mutation absent, and NPM1 mutation present, no significant differences were observed among any of these characteristics (P> 0.05).
In the entire cohort, no significant differences were observed in EFS or OS of patients between the DNMT3AmutIDH1/2wild and DNMT3AmutIDH1/2mut groups (Figure 2A and B, P> 0.05). We divided the patients into 2 groups according to treatment, and observed no significant differences in either group (Figure 2C–F, P> 0.05). The HSCT showed better efficacy than chemotherapy in the DNMT3AmutIDH1/2wild group, and led to a higher EFS and OS (Figure 3A and B, P=0.010 for EFS and P=0.007 for OS). However, in the DNMT3AmutIDH1/2mut group, no significant differences were observed between the two treatment methods (Figure 3C and D, P> 0.05).
Considering the differences in effect of IDH1 and IDH2 mutations on the prognosis of patients, we divided the entire cohort into 3 groups, more specifically the IDH1/2wild, IDH1 mutation and IDH2 mutation groups. We did not separate IDH2R140 and IDH2R172 mutations because of the small sample size. The results of Kaplan–Meier analysis for EFS and OS are shown in Figure 4A–D (P> 0.05). No significant differences were observed among the 3 groups.
We further verified our results in the Clinseq cohort. When patients with mutations of IDH1 or IDH2 were grouped together, the results of Kaplan–Meier analysis for OS showed no significant differences between the DNMT3AmutIDH1/2wild and DNMT3AmutIDH1/2mut groups (Figure 5A, P=0.128). In Figure 5B, patients with the IDH2R140 mutation were considered a single group. Furthermore, because of their relatively consistent effects on the prognosis, patients with mutations of IDH1 and IDH2R172 were divided into 2 groups. No significant differences were observed in the OS among these 3 groups (P=0.124).
Discussion
We synthetically analyzed different aspects of the effects of IDH1/2mut on the outcomes of patients with DNMT3A-mutated AML; however, no significant differences were found. The HSCT could be a more favorable option for patients with the DNMT3A mutation. However, HSCT and chemotherapy showed no significant differences in patients with double mutations. The DNMT3AmutIDH1/2mut group showed relatively lower rates of NPM1 mutation.
Previous studies have indicated that NPM1 and double-mutated CEBPA have positive effects, whereas FLT3-ITD, TET2, KRAS, PTPN11, U2AF1, and MLL-PTD mutations have adverse effects on the outcomes of AML patients.20–27 Mutations of SMC3, RAD21, and NRAS were thought to have little to no influence on those studies, whereas the effects of other gene mutations, such as that of SMC1A, on the prognosis of AML had not been clearly determined.20–27
In our cohort, 54.9% of the patients had NPM1 mutations. Patients in the DNMT3AmutIDH1/2mut group had a lower mutation rate of NPM1 (45.1% versus 9.8%). In another study, about 80% of DNMT3A-mutated patients had NPM1 mutations, and 65.4% of the double-mutation patients concurrently harbored NPM1 mutations.8 Rakheja et al indicated that IDH1R132 and IDH2R140 mutations are frequently accompanied by NPM1 mutations in AML.10 In our cohort, 87.5% of the patients harbored IDH1R132 or IDH2R140; however, we observed no correlation between NPM1 and IDH1/2. The reason for this difference might be due to the age distribution of the sample, the small sample size, or the research background of our cohort.
Many studies have attempted to elucidate the effects of multiple gene mutations on the outcomes of AML. More specifically, studies have shown that FLT3-ITD with the DNMT3AR882 double mutation is a poor prognostic factor in AML;28 whereas NPM1 mutations do not seem to have a significant effect on the outcomes of patients with DNMT3A-mutated AML.8 Alternatively, IDH1/2mut constitute a poor prognostic factor in NPM1-mutated cytogenetically normal-AML (CN-AML) without FLT3-ITD.29 Our study revealed that IDH1/2mut have little influence on the outcomes of patients with DNMT3A-mutated AML.
The HSCT has been confirmed as an effective therapy for intermediate- or high-risk patients with AML. Patients at higher risk usually benefit more from transplant therapy. The survival of CN-AML patients with DNMT3Amut could be improved following allo-HSCT.30 Nevertheless, as we have mentioned before, the FLT3-ITD and DNMT3AR882 double mutation is a poor prognostic factor in AML. Patients with this condition have significantly lower 2-year OS and leukemia-free survival (the duration from post-transplantation to recurrence of leukemia or death) following allo-HSCT, than patients with single FLT3-ITD or DNMT3Amut.28
In comparison with chemotherapy, our results demonstrate that HSCT had a more favorable therapeutic effect on patients with the DNMT3A mutation. When IDH1/2mut were taken into account however, the effects of these two therapies were statistically equivalent. Patients with the DNMT3A and IDH1/2 double mutation showed no gain in EFS or OS after either HSCT or chemotherapy. This result suggests that IDH1/2mut might not be a strong biomarker for HSCT therapy in patients with the DNMT3A mutation. Analysis of data from the Clinseq cohort further supported the view that IDH1/2mut in patients with AML who received chemotherapy had little effect on the outcomes of those with the DNMT3A mutation. The results of the analysis in which different mutations of IDH were separated also showed that they had no impact on the prognosis of these patients, even though mutations of IDH1, IDH2R140, and IDH2R172 might have different effects on the outcome.
In addition to HSCT, targeted therapy is now one of the more popular treatment choices that can show good efficacy. Enasidenib (AG-221) is a reversible and selective inhibitor of mutant IDH2.3,31,32 Inhibitors of IDH1, such as AG-120 and IDH305, are also under investigation.3 Although our study showed that IDH1/2mut have no influence on the outcomes of AML patients with DNMT3Amut, because of the development of novel, effective target drugs, IDH1/2 inhibitors might have positive effects on the overall prognosis of patients with double mutations.
Limitations
Several limitations of the present study need to be acknowledged. Our study is a retrospective study with a relatively small sample size, and some bias needs to be considered. In some groups, the number of samples was less than 5, thus limiting further statistical analysis. Therefore, the present results need to be verified in larger cohorts.
Conclusion
We propose that IDH1/2 has no impact on the prognosis of AML patients with the DNMT3A mutation. Furthermore, HSCT showed a more favorable therapeutic effect for patients with the DNMT3A, but without the IDH1/2 mutation. For patients with double mutations, the efficacy of HSCT was similar to that of chemotherapy.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81500118, 61501519), the China Postdoctoral Science Foundation funded project (project no. 2016M600443), and the PLAGH project of Medical Big Data (project no. 2016MBD-025). Xinpei Zhang and Jinlong Shi contributed equally as co-first authors.
Author contributions
Xiaoyan Ke and Lin Fu designed the study; Xinpei Zhang wrote the manuscript; Xinpei Zhang, Jinlong Shi, Jilei Zhang, Xinrui Yang, Gaoqi Zhang, Siyuan Yang, and Jing Wang performed statistical analyses and analyzed the data. Xiaoyan Ke and Lin Fu coordinated the study over the entire experimental period. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. | ||
Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. | ||
Kavanagh S, Murphy T, Law A, et al. Emerging therapies for acute myeloid leukemia: translating biology into the clinic. JCI Insight. 2017;2(18):e95679. | ||
Sehgal AR, Gimotty PA, Zhao J, et al. DNMT3A mutational status affects the results of dose-escalated induction therapy in acute myelogenous leukemia. Clin Cancer Res. 2015;21(7):1614–1620. | ||
Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. | ||
Hájková H, Marková J, Haškovec C, et al. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk Res. 2012;36(9):1128–1133. | ||
Thol F, Damm F, Lüdeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896. | ||
Gale RE, Lamb K, Allen C, et al. Simpson’s paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemia. J Clin Oncol. 2015;33(18):2072–2083. | ||
Hou HA, Kuo YY, Liu CY, et al. DNMT3A mutations in acute myeloid leukemia: Stability during disease evolution and clinical implications. Blood. 2012;119(2):559–568. | ||
Rakheja D, Konoplev S, Medeiros LJ, Chen W. IDH mutations in acute myeloid leukemia. Hum Pathol. 2012;43(10):1541–1551. | ||
DiNardo CD, Ravandi F, Agresta S, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90(8):732–736. | ||
Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. | ||
Im AP, Sehgal AR, Carroll MP, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28(9):1774–1783. | ||
Upadhyay VA, Brunner AM, Fathi AT. Isocitrate dehydrogenase (IDH) inhibition as treatment of myeloid malignancies: Progress and future directions. Pharmacol Ther. 2017;177:123–128. | ||
Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. | ||
Xu Q, Li Y, Lv N, et al. Correlation between isocitrate dehydrogenase gene aberrations and prognosis of patients with acute myeloid leukemia: A systematic review and meta-analysis. Clin Cancer Res. 2017;23(15):4511–4522. | ||
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. | ||
Wang M, Lindberg J, Klevebring D, et al. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31(10):2029–2036. | ||
Wahlin A, Billström R, Björ O, et al. Results of risk-adapted therapy in acute myeloid leukaemia. A long-term population-based follow-up study. Eur J Haematol. 2009;83(2):99–107. | ||
Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. | ||
Zhou J-D, Yao D-M, Li X-X, et al. KRAS overexpression independent of RAS mutations confers an adverse prognosis in cytogenetically normal acute myeloid leukemia. Oncotarget. 2017;8(39):66087–66097. | ||
Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: A study of 2502 patients. Blood. 2006;107(10):3847–3853. | ||
Kadia TM, Kantarjian H, Kornblau S, et al. Clinical and proteomic characterization of acute myeloid leukemia with mutated RAS. Cancer. 2012;118(22):5550–5559. | ||
Thol F, Bollin R, Gehlhaar M, et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123(6):914–920. | ||
Andrade FG, Noronha EP, Brisson GD, et al. Molecular characterization of pediatric acute myeloid leukemia: Results of a multicentric study in Brazil. Arch Med Res. 2016;47(8):656–667. | ||
Ohgami RS, Ma L, Merker JD, et al. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol. 2015;28(5):706–714. | ||
Sun QY, Ding LW, Tan KT, et al. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD). Leukemia. 2017;31(1):1–10. | ||
Tang S, Shen H, Mao X, et al. FLT3-ITD with DNMT3A R882 double mutation is a poor prognostic factor in Chinese patients with acute myeloid leukemia after chemotherapy or allogeneic hematopoietic stem cell transplantation. Int J Hematol. 2017;106(4):552–561. | ||
Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. | ||
Xu Y, Sun Y, Shen H, et al. Allogeneic hematopoietic stem cell transplantation could improve survival of cytogenetically normal adult acute myeloid leukemia patients with DNMT3A mutations. Am J Hematol. 2015;90(11):992–997. | ||
Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722–731. | ||
Amatangelo MD, Quek L, Shih A, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 2017;130(6):732–741. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.