Back to Journals » International Journal of General Medicine » Volume 15
CLEC1B is a Promising Prognostic Biomarker and Correlated with Immune Infiltration in Hepatocellular Carcinoma
Authors Liang X , Song F, Fang W , Zhang Y, Feng Z, Chen Z , Han L, Chen Z
Received 18 February 2022
Accepted for publication 10 May 2022
Published 16 June 2022 Volume 2022:15 Pages 5661—5672
DOI https://doi.org/10.2147/IJGM.S363050
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xiaoliang Liang,1,* Fei Song,2,* Wanzhi Fang,1 Yu Zhang,1 Zihan Feng,1 Zeyin Chen,1 Lu Han,3 Zhong Chen1
1Department of Hepatobiliary Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, People’s Republic of China; 2Department of General Surgery, Yancheng TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Yancheng, 224002, People’s Republic of China; 3Department of Medicine, Jiangsu Vocational College of Medicine, Yancheng, 224005, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhong Chen, Department of Hepatobiliary Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, People’s Republic of China, Email [email protected]
Purpose: C-type lectin domain family 1 member B (CLEC1B) is a protein-coding gene involved in various processes, such as platelet activation, tumor cell metastasis and separation of blood/lymphatic vessels. However, how CLEC1B plays its role in hepatocellular carcinoma (HCC) has not been well studied. The purpose of this study was to investigate the clinical significance and biological function of CLEC1B in HCC.
Patients and Methods: Based on (The Cancer Genome Atlas) TCGA database, CLEC1B expression matrix and corresponding clinical information were extracted. ROC curves and Kaplan–Meier method were generated to evaluate the value of CLEC1B as a diagnostic and prognostic biomarker. Moreover, single-gene difference analysis constructed by DESeq2 method and then the related genes were used to predict CLEC1B-related signaling pathways. The ssGSEA algorithm was conducted for studies related to immune infiltration. CLEC1B protein expression was evaluated and immunohistochemistry in HCC tissues through tissue microarray. Finally, the relationship between CLEC1B expression and T cell infiltration was assessed according to tissue microarray.
Results: The mRNA and protein levels of CLEC1B were significantly down-regulated in HCC compared to paired normal tissues, which were further verified in clinical tissue samples. ROC curves and Kaplan–Meier survival analysis suggested the significant diagnostic and clinical prognostic value of CLEC1B. Meanwhile, downregulation of CLEC1B was significantly associated with clinical parameters such as clinical tumor vascular invasion and distant metastasis. Moreover, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene set enrichment (GSEA) analysis indicated that CLEC1B has significant association with immune function. Finally, immune infiltration analysis indicated that CLEC1B was significantly associated with immune cell subsets and affected the efficacy of immunotherapy in cancer patient.
Conclusion: Collectively, our findings suggested that CLEC1B could be a promising prognostic biomarker in HCC and its expression was related to immune cell infiltration.
Keywords: CLEC1B, hepatocellular carcinoma, prognostic value, tumor immune infiltration, bioinformatics analysis
Introduction
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer-associated mortality globally, accounting for 75–85% of primary liver cancer case and its incidence is on the rise.1,2 Cirrhosis, aflatoxin contamination and heavy drinking are major risk factors for HCC.3 Although early-stage HCC can often be treated with potentially curative partial resection or liver transplantation or with locoregional procedures, which offer the best 5-year survival rates, at 47.9% and 59.3%, respectively.4,5 Because, there are no specific symptoms at the early stage, HCC is usually diagnosed in advanced stages, resulting in limited treatment options and survival rates among HCC patients also remain challenging in the clinical practice.6 It is essential to determine the etiology and mechanisms of HCC malignant progression and explore more effective treatment strategies.
In recent years, there is emerging data suggesting targeting the host immune system can enhance cancer therapies.7 Cancer immunotherapy is revolutionizing the clinical management of a broad range of solid tumor types.8 Unfortunately, the PD-1/PDL-1 inhibitor, known as immune checkpoint blockade (ICB) with the best responses, is limited to subsets of patients, with overall response rates of 20% or less.8–10 The tumor microenvironment (TME) is an essential component of the tumor structure, which is associated with ICB responsiveness.11,12 An increasing number of researches on TME have revealed a crucial role of the tumor-infiltrating immune cells in tumor relapse, metastasis, and therapeutic response to immunotherapy.13,14 Typically, the low infiltration of CD8+T cells and a high proportion of tumor-associated macrophages (TAMs) result in tumor growth, immune evasion and poor prognosis of patients, which is associated with poor clinical outcomes in most carcinoma cases.15,16 To increase the efficacy and durability of immunotherapy, a new marker correlated with the landscape of TME and a better understanding of immunotherapy resistance mechanisms are needed.
CLEC1B is a type II transmembrane receptor of the C-type lectin superfamily, which is characterized by one or more C-type lectin-like domains (CTLDs).17 CLEC1B is a platelet-related molecule that activating receptor for the snake venom toxin rhodocytin as well as endogenous ligand podoplanin, and also plays crucial roles in lymphatic/blood vessel separation, tumor cell-induced platelet aggregation and immune response.18–20 Previous studies have been reported that CLEC1B have an inhibitory effect on platelet aggregation and tumor metastasis in colon carcinoma.21 Moreover, Wang Lan et al and WangYing et al found that CLEC1B suppresses metastasis of gastric cancer cells by preventing activation of AKT and GSK3B signaling.20,22 Recently, low CLEC1B and high PD-L1 have been reported to be a valuable prognostic factor implying worse clinical outcomes in HCC.23 However, the role of CLEC1B in the tumor immune microenvironment and progression of hepatocellular carcinoma has not been fully elucidated.
In this study, we aimed to explore comprehensively and systematically explore the expression of CLEC1B in HCC through bioinformatics analysis and clinical tissue microarray samples. Our present study sheds light on the critical role of CLEC1B in improved cancer survival and immune infiltration in HCC. Therefore, CLEC1B could serve as a new potential immune therapeutic and prognostic biomarker for HCC.
Materials and Methods
Patients and Specimens
Tumor specimens were obtained from 68 consecutive HCC patients who received liver resection in the Liver Cancer Institute, the Affiliated Hospital of Nantong University from February 2018 to December 2019. All cases were re-examined by experienced histopathologists to confirm the diagnosis. Histopathological diagnosis was made based on World Health Organization criteria, and tumor staging was defined according to the Seventh Edition of Tumor-Node-Metastasis (TNM) Classification of International Union against Cancer. Clinical data collection, postoperative follow-up procedures and admission criteria were performed according to the unified guidelines described in our previous study. Ethical approval for human subjects was obtained from the research ethics committee of Affiliated Hospital of Nantong University, and informed consent was obtained from each patient. Clinical samples were collected and processed in accordance with the Declaration of Helsinki.
TCGA and GEO Database
Transcriptional expression data of CLEC1B and corresponding clinical information were downloaded from TCGA and GEO website. The data from TCGA were comprised of 374 LIHC samples and 50 paracancerous tissues. The bioinformatic analysis about CLEC1B was achieved from TCGA database. The data from GEO include 110 normal liver tissues. The gene expression data with workflow type of FPKM was transformed into TPM format and log2 conversion for further study. For pan-cancer analysis, normal RNA-Seq data for 33 kinds of tumors were obtained from TCGA. Result was achieved from TCGA database. The Wilcoxon rank sum test detected two sets of data, and p < 0.05 was considered statistically significant (ns, p > 0.05; *, p < 0.05; **p < 0.01; ***, p < 0.001).
Functional Enrichment Analysis
To identify CLEC1B potential function, Gene Ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed by clusterProfiler R package. P-value <0.05 indicated significant GO terms and KEGG pathways. Gene set enrichment analysis (GSEA) uses genome-wide expression profiling microarray data to compare gene enrichment to a predefined gene set (21). Gene expression data were divided into two groups according to CLEC1B expression level: CLEC1B high and CLEC1B low. The dataset used for GSEA analysis is the molecular signatures database (MsigDB), where the hallmark gene set was used in this analysis. Significant enrichment was defined as a gene set with a standard: |NES| > 1, p-value <0.05, FDR < 0.25.
Analysis of Tumor-Infiltrating Immune Cells
The assessment of tumor-infiltrating immune cells was performed using the ssGSEA algorithm. Briefly, ssGSEA algorithm is a tool that can be used to evaluate the infiltration of various immune cells and their immune-related signature using GSVA R package. Besides, based on the differentially expressed gene markers of CLEC1B gene expression profile, the immune infiltration score, matrix score, and estimated score were calculated for the samples. The correlations between CLEC1B and TILs were measured by Spearman’s test. The Wilcoxon rank sum test detected two sets of data: CLEC1B high and CLEC1B low.
TMA Construction and Immunohistochemistry
The human HCC tissue microarray was obtained from Nantong University including 68 HCC tissue samples and paired non-tumor tissue samples. IHC staining was carried out to measure CLEC1B protein level in HCC tissues per the instructions from the manufacturer using an anti-CLEC1B, CD4, CD8 antibody (1:1000, Proteintech). Each HCC sample was evaluated based on the staining intensity and the percentage of cells with positive staining. Immunostaining intensities of these markers were semi-quantitatively scored as follows: 0–1, negative; 2–3, weak; 4–5, moderate; 6–8, strong. The score of immunostaining intensity was assessed by two pathologists independently, and comparisons were performed between tumor/normal samples.
Diagnostic Value Analysis and Survival Prognosis Analysis
The diagnostic value of CLEC1B in HCC was assessed by receiver operating characteristic (ROC) curve. The area under the ROC curve (AUC) indicated the magnitude of diagnostic efficiency. The closer the area under the curve (AUC) is, the better the diagnostic effects. AUC in 0.5–0.7 has a low accuracy, AUC in 0.7–0.9 has a certain accuracy, and AUC above 0.9 has a high accuracy, respectively. Kaplan–Meier plots were employed to assess the relationship between CLEC1B expression and prognosis (OS, DSS, and PFI) of cancers.
Statistical Analysis
All statistical analyses were performed using the SPSS software and the GraphPad Prism software was used for graphing the histogram and analyze the different expression between adjacent tissues and tumor tissues of IHC staining. Quantitative values are presented as mean ± SD or median (range). Pearson Χ2 test was performed to evaluate the independent correlation between CLEC1B expression level and CD8+ and CD4+ T cell infiltration in HCC tissue. Spearman correlation coefficient (r) was used to access the correlation between lymphocyte infiltration and CLEC1B expression. The GO and KEGG enrichment analyses were visualized using the R packages “ggplot2” and “cluster Profiler”, the threshold for statistically significant differences was set at P adjust ≤0.05. In addition, all P values were adopted by a two-sided test and P < 0.05 was regarded as having statistical significance.
Results
CLEC1B is Down-Regulated in Hepatocellular Carcinoma and Multiple Human Cancers
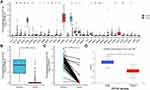
We examined CLEC1B expression levels in different types of cancer using the Cancer Genome Atlas (TCGA) database, the transcriptional levels of CLEC1B were significantly downregulated in various human cancer tissue compared to normal tissue (Figure 1A). For further assessments about the mRNA and protein expression of CLEC1B in hepatocellular carcinoma, we retrieved the differential expression of CLEC1B in cancer vs adjacent carcinoma in individual tumor samples from the TCGA, GEO and Clinical Proteomic Tumor Analysis Consortium (CPTAC) databases. Unpaired and paired data analysis indicated that the mRNA expression levels of CLEC1B in hepatocellular carcinoma tissues significantly decreased in HCC samples relative to normal liver samples (Figure 1B). This conclusion was also verified in paired HCC and normal tissues (Figure 1C). The data from CPTAC also showed that CLEC1B protein was downregulated in hepatocellular carcinoma tissue. From the expression analysis of CLEC1B in hepatocellular carcinoma, it could be found that the expression level of CLEC1B is generally lower in tumors than that in normal tissues.
Low CLEC1B Expression is Correlated with Poor OS in HCC Patients and May Serve as a Diagnostic Biomarker Based on TCGA Datasets
To investigate the value of CLEC1B to distinguish HCC samples from normal samples, we performed a ROC curve analysis. The ROC curve analysis demonstrated the strong value of CLEC1B in the diagnosis of HCC: 0.986 (95% CI: 0.976–0.996) for CLEC1B (Figure 2A). To explore the association between CLEC1B level and prognosis in HCC patients, we used survival analysis and found that the Overall Survival (OS), Relapse Free Survival (RFS), Progression Free Survival (PFS) and Disease Specific Survival (DSS) rates of HCC patients with low CLEC1B expression were significantly lower than those of patients with high CLEC1B expression (Figure 2B and C). These results implied that high expression of CLEC1B is a potential prognostic indicator molecule in hepatocellular carcinoma.
Low CLEC1B Expression Correlates with Aggressive Clinical Parameters
To explore whether CLEC1B mRNA expression is associated with clinical implications in hepatocellular carcinoma, we performed Mann–Whitney U-tests and logistic regression analyses. A negative trend was found between expression of CLEC1B in HCC and the progression of tumor distant metastasis (Figure 3A–N). The analysis of additional parameters found low CLEC1B expression was significantly correlated with aggressive tumor characteristics, such as tumor metastasis stage (P < 0.05) and vascular invasion (P < 0.01). In conclusion, these results suggested that low CLEC1B may act as a biomarker for metastasis in hepatocellular carcinoma.
Functional Enrichment Analysis of Differentially Expressed Genes Between CLEC1B-High and CLEC1B-Low Group in HCC
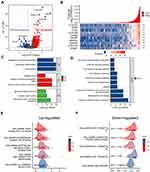
To explore the specific role of CLEC1B protein and downstream pathways caused by high expression of CLEC1B. We constructed a volcano plot to elucidate the differentially expressed genes (DEGs) between HCC samples between CLEC1B high and CLEC1B low mRNA group in HCC based on the TCGA data (Figure 4A). There are 837 upregulated protein-coding genes and 109 protein-coding genes and heatmaps depicted the top 10 significantly upregulated DEGs between the CLEC1B high and CLEC1B low expression groups (Figure 4B). Besides, we performed GO enrichment analysis revealed that the biological processes of CLEC1B are mainly associated with T cell activation and lymphocyte differentiation (Figure 4C). The results of KEGG enrichment also revealed the enrichment of T cell receptor signaling pathway, which corresponded to the results of the GO enrichment analysis (Figure 4D). The CLEC1B-related genes were further analyzed using GSEA to identify signaling pathways that were significantly enriched in HCC. Intriguingly, GSEA analysis of differentially expressed genes revealed three of the top five most positively regulated hallmark signatures were inflammatory response, interferon gamma response and TNFα signaling (Figure 4E). Moreover, the results of the downregulated signaling pathway in the GSEA enrichment analysis showed its negative association with tumor progression: DNA repair, E2F target and MYC target (Figure 4F). Collectively, these findings suggest that CLEC1B activates immune response signaling in HCC leading to secretion of essential pro-inflammatory cytokines, which in turn can activate CD8+ T cells.
High Expression of CLEC1B Was Associated with Low CD8 + T Cell Levels Could Predict the Clinical Benefit of ICB
Considering the fact that inflammatory response and infiltrating immune cells can affect the prognosis of HCC, we also evaluated the relationship between CLEC1B expression and immune cell infiltration using the ssGSEA algorithm based on TCGA data. We demonstrated that CLEC1B expression was positively correlated with the abundance of immunocytes such as B cells, Macrophages, NK cells, T cells, T helper 1 (Th1) cells and cytotoxic cells (Figure 5A and B, Figure S1). Next, to explore the roles that the CLEC1B influenced tumor immune microenvironment has during tumor development, we estimate immune score, microenvironment score and stroma score using the R package estimate (Figure 5C). They significantly increased in higher group. In addition, a comprehensive heatmap illustrates the differences of immune checkpoints, cytolytic activity signature, and IFN-γ response signature between the CLEC1B high and CLEC1B low expression clusters (Figure 5D). These data indicated that CLEC1B may play a specific role in immune infiltration and a potential predictor about immunotherapy response of hepatocellular carcinoma.
Low Expression of CLEC1B Was Associated with Suppressive Immune Microenvironment
The study further explored the CLEC1B expression and its clinical relevance with CD8+ and CD4+ T cells in HCC samples by Tissue Microarray analysis (TMA) immunohistochemical staining. In comparison with normal paracancerous tissues, the staining data showed that CLEC1B significantly reduced in HCC tissue: strong expression (30.9%), moderate expression (20.6%), weak expression (29.4%) and negative expression (19.1%). In contrast, in normal liver tissue, CLEC1B expression was shown as follows: strong expression (51.5%), moderate expression (19.1%), weak expression (25%) and negative expression (4.4%) (Figure 6A). In order to verify the clinical significance of CLEC1B and its relationship between the tumor-infiltrating lymphocytes in HCC, all 68 HCC patients were divided into CLEC1B negative (score negative, n = 13) and CLEC1B positive (score weak, moderate or strong, n = 55) groups based on immunohistochemical data. As expected, the expression of CLEC1B was positively associated with the CD8+ and CD4+ T cells infiltration level (P < 0.05) (Figure 6B). The presence of CD8+T cell and CD4+T cells in the tumor microenvironment are also critical of antitumor immunity and better response with immune checkpoint inhibitor. In summary, the results of the patient samples suggested that low CLEC1B expression in HCC tissues and its correlation with T cell infiltration, which supported low CLEC1B expression could be a biomarker for the immune suppressive microenvironment.
Discussion
HCC is one of the worldwide life-threatening malignant tumors, which is in urgent need for expanding the understanding of underlying mechanism to support therapy development.24 Many factors, such as activation of oncogenes and inactivation of tumor suppressor genes, could cause unrestricted growth of tumor cells.25–27 Among them, hundreds of cancer-associated genes are closely related to the occurrence and development of HCC.28 With the development of a variety of “omics” technologies such as genomics, proteomics and transcriptomics, accumulating molecular markers related to malignant transformation of HCC cells have been discovered.29,30 At the same time, with the understanding and acquaintance of tumor immunobiology deepening among researchers, the breakthroughs in the field of tumor immunotherapy in recent years have provided new approaches for cancer therapy.31,32 Immunotherapy has transformed the treatment of many advanced malignant tumors.33,34 Therefore, further study of HCC-related genes and identifying novel treatment targets that predict the response rate of immunotherapy better has become an urgent task.
CLEC1B is responsible for platelet activation, tumor cell metastasis, separation of blood/lymphatic vessels and cerebrovascular patterning during embryonic development.17 In previous study, CLEC1B is proved to have an inhibitory effect on platelet aggregation and tumor metastasis by binding to the surface of tumor cells, which is associated highly with spontaneous tumor hemorrhage (TH).23,35 There is emerging evidence supported the strong association between TH and tumor microenvironment.23,36 For instance, TH in HCC implies tumor microenvironment of red blood cells releasing and platelet aggregation, which could promote tumor growth, invasion, and metastasis partially through the activation of NF-kB pathway.37,38 However, the role of CLEC1B in HCC progression and tumor microenvironment as well as its potential mechanism remains poorly understood.
In this study, we assessed CLEC1B expression using RNA-seq data from multiple malignancies in the TCGA database. Through the GO and KEGG analysis of the biological role of CLEC1B, we found that CLEC1B performed a crucial role in inflammatory activities and immunological reactions in LIHC. Our TCGA analysis also confirmed that low CLEC1B expression levels are associated with poor survival in the hepatocellular carcinoma cohort. Finally, we validated our resulting molecular trends in clinical specimens, and immunohistochemistry findings revealed that the low CLEC1B expression group was associated with less CD8+ T cells and CD4+ T cells infiltration compared to those with high CLEC1B protein expression group. Notably, low protein expression of CLEC1B was associated with more progressive clinical features such as tumor status, vascular invasion and metastasis. Overall, these findings provide strong support for the use of CLEC1B expression as a diagnostic and prognostic biomarker for HCC.
The immune responses at tumor sites are determined by the TME-infiltrating immune cells. The association between CLEC1B and antitumour immunity may be related to multiple mechanisms. Further, ssGSEA method was used to explore the correlation of CLEC1B expression with immune cell infiltration degrees in tumors, which suggested that CLEC1B expression was positively correlated with the infiltration degree of B cells, NK cells, T cells, macrophages, Th1 cells and cytotoxic T cells in HCC. We also found that CLEC1B expression was significantly correlated with three immune response signalings in HCC, which may indicate its role in the development and application of certain drugs in the future. In general, the relationship of CLEC1B expression with immune cell markers revealed that CLEC1B was involved in regulating tumor immunity in HCC. These findings provide strong evidence that CLEC1B is involved in tumor immune infiltration.
Many studies have shown that in the process of the occurrence and development of liver cancer, tumors often form an immunosuppressive microenvironment favorable to their own growth through ways such as secreting inhibitory cytokines and expressing immune checkpoints, thereby achieving immune escape.39 Even though immune checkpoint inhibitors, represented by PD-1/PD-L1, are used to activate anti-tumor immunity, immunotherapy is often unsatisfactory due to its low efficiency in population. The mechanism of resistance of tumor immune checkpoint inhibitors is mainly due to the low expression of PD-L1, the lack of T cells in the cold tumor environment (which lacks of T cells in the tumor microenvironment), and the inhibition of interferon gamma (IFNG) signaling pathways.40 Although CLEC1B has been reported as an indicator for predicting spontaneous tumor hemorrhage in HCC, further research is needed on the association of this protein in the regulation of the immune microenvironment.23 When we explored the association of CLEC1B with the immune microenvironment, CLEC1B shows positive correlation for a variety of immune-infiltrating cells, especially T, B, and NK cells. Its bioinformatics analysis shows that the molecule is associated with anti-tumor immunity as well. These results imply that the high expression of this protein can significantly increase the number of T cells in HCC, realize the conversion of cold to hot tumor environment in HCC, so as to realize the sensitization of immune checkpoint inhibitors. Although the association between CLEC1B and tumor infiltrating immune cells was observed, how CLEC1B triggers antitumor immune cell infiltration is still unsolved and need to be further clarified.
Conclusion
In summary, our findings indicate that CLEC1B may serve as a promising prognostic biomarker and its low expression correlated with immunosuppression in hepatocellular carcinoma. CLEC1B is often down-regulated in HCC and inhibits tumor progression by activating antitumor immunity.
Abbreviations
CLEC1B, C-type Lectin Domain Family 1 Member B; HCC, Hepatocellular Carcinoma; TCGA, The Cancer Genome Atlas; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment Analyses; OS, Overall Survival; RFS, Relapse Free Survival; PFS, Progression Free Survival; DSS, Disease Specific Survival; TMA, Tissue Microarray; TME, tumor microenvironment; ROC, receiver operating characteristic.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (grant no. 81871927).
Author Contributions
All authors contributed significantly to the reported work, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; were involved in the drafting, review and revision of the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Xiaoliang Liang and Fei Song contributed equally to this work as first authors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Dawkins J, Webster RM. The hepatocellular carcinoma market. Nat Rev Drug Discov. 2019;18:13–14. doi:10.1038/nrd.2018.146
2. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi:10.1038/s41575-019-0186-y
3. Bresnahan E, Lindblad KE, Ruiz de Galarreta M, Lujambio A. Mouse models of oncoimmunology in hepatocellular carcinoma. Clin Cancer Res. 2020;26:5276–5286. doi:10.1158/1078-0432.CCR-19-2923
4. Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi:10.1038/nrdp.2016.18
5. Fleming BD, Urban DJ, Hall MD, et al. Engineered Anti-GPC3 Immunotoxin, HN3-ABD-T20, produces regression in mouse liver cancer xenografts through prolonged serum retention. Hepatology. 2020;71:1696–1711. doi:10.1002/hep.30949
6. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi:10.1016/S0140-6736(18)30010-2
7. Holtzhausen A, Harris W, Ubil E, et al. TAM family receptor kinase inhibition reverses mdsc-mediated suppression and augments Anti-PD-1 therapy in melanoma. Cancer Immunol Res. 2019;7:1672–1686. doi:10.1158/2326-6066.CIR-19-0008
8. Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, et al. β-catenin activation promotes immune escape and resistance to Anti-PD-1 Therapy in hepatocellular carcinoma. Cancer Discov. 2019;9:1124–1141. doi:10.1158/2159-8290.CD-19-0074
9.. Ott PA, Hodi FS, Kaufman HL, Wigginton JM, Wolchok JD. Combination immunotherapy: a road map. J Immunother Cancer. 2017;5. doi:10.1186/s40425-017-0218-5
10. Finn R, Ryoo B-Y, Merle P, et al. Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE-240: a randomized, double-blind, phase III trial. J Clin Oncol. 2020;38:193–202. doi:10.1200/jco.19.01307
11. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi:10.1038/s41591-018-0014-x
12. de Azevedo RA, Shoshan E, Whang S, et al. MIF inhibition as a strategy for overcoming resistance to immune checkpoint blockade therapy in melanoma. Oncoimmunology. 2020;9. doi:10.1080/2162402X.2020.1846915
13. Jiang Y, Zhang Q, Hu Y, et al. ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg. 2018;267:504–513. doi:10.1097/SLA.0000000000002116
14. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17:807–821. doi:10.1038/s41423-020-0488-6
15. Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi:10.1038/nature22079
16. Feng Q, Chang W, Mao Y, et al. Tumor-associated macrophages as prognostic and predictive biomarkers for postoperative adjuvant chemotherapy in patients with stage II colon cancer. Clin Cancer Res. 2019;25:3896–3907. doi:10.1158/1078-0432.CCR-18-2076
17. Meng D, Luo M, Liu B. The role of CLEC-2 and its ligands in thromboinflammation. Front Immunol. 2021;12. doi:10.3389/fimmu.2021.688643
18. Ozaki Y, Tamura S, Suzuki-Inoue K. New horizon in platelet function: with special reference to a recently-found molecule, CLEC-2. Thromb J. 2016;14. doi:10.1186/s12959-016-0099-8
19. Suzuki-Inoue K, Osada M, Ozaki Y. Physiologic and pathophysiologic roles of interaction between C-type lectin-like receptor 2 and podoplanin: partners from in utero to adulthood. J Thromb Haemost. 2017;15:219–229. doi:10.1111/jth.13590
20. Wang L, Yin J, Wang X, et al. C-type lectin-like receptor 2 suppresses AKT signaling and invasive activities of gastric cancer cells by blocking expression of phosphoinositide 3-kinase subunits. Gastroenterology. 2016;150:1183–1195.
21. Suzuki-Inoue K, Kato Y, Inoue O, et al. Involvement of the snake toxin receptor CLEC-2, in podoplanin-mediated platelet activation, by cancer cells. J Biol Chem. 2007;282:25993–26001. doi:10.1074/jbc.M702327200
22. Wang Y, Lv Y, Liu TS, et al. Cordycepin suppresses cell proliferation and migration by targeting CLEC2 in human gastric cancer cells via Akt signaling pathway. Life Sci. 2019;223:110–119.
23. Hu K, Wang Z-M, Li J-N, et al. CLEC1B expression and PD-L1 expression predict clinical outcome in hepatocellular carcinoma with tumor hemorrhage. Transl Oncol. 2018;11:552–558. doi:10.1016/j.tranon.2018.02.010
24. Song H, Liu Y, Li X, et al. Long noncoding RNA CASC11 promotes hepatocarcinogenesis and HCC progression through EIF4A3-mediated E2F1 activation. Clin Transl Med. 2020;10. doi:10.1002/ctm2.220
25. Zeineldin M, Federico S, Chen X, et al. MYCN amplification and ATRX mutations are incompatible in neuroblastoma. Nat Commun. 2020;11. doi:10.1038/s41467-020-14682-6
26. Rehmani H, Li Y, Li T, et al. Addiction to protein kinase Cɩ due to PRKCI gene amplification can be exploited for an aptamer-based targeted therapy in ovarian cancer. Signal Transduct Target Ther. 2020;5. doi:10.1038/s41392-020-0197-8
27. Molinie N, Rubtsova SN, Fokin A, et al. Cortical branched actin determines cell cycle progression. Cell Res. 2019;29:432–445. doi:10.1038/s41422-019-0160-9
28. Xu Q, Li Y, Gao X, et al. HNF4α regulates sulfur amino acid metabolism and confers sensitivity to methionine restriction in liver cancer. Nat Commun. 2020;11:1–7. doi:10.1038/s41467-020-17818-w
29. Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi:10.1053/j.gastro.2016.11.048
30. Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12. doi:10.1186/s13045-019-0806-6
31. Ma W, Gilligan BM, Yuan J, Li T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol. 2016;9:47. doi:10.1186/s13045-016-0277-y
32. Kennedy LB, Salama AKS. A review of cancer immunotherapy toxicity. CA Cancer J Clin. 2020;70:86–104. doi:10.3322/caac.21596
33. Rosato PC, Wijeyesinghe S, Stolley JM, et al. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat Commun. 2019;10. doi:10.1038/s41467-019-08534-1
34. Waks AG, Stover DG, Guerriero JL, et al. The immune microenvironment in hormone receptor-positive breast cancer before and after preoperative chemotherapy. Clin Cancer Res. 2019;25:4644–4655. doi:10.1158/1078-0432.CCR-19-0173
35. Tsukiji N, Inoue O, Morimoto M, et al. Platelets play an essential role in murine lung development through Clec-2/podoplanin interaction. Blood. 2018;132:1167–1179. doi:10.1182/blood-2017-12-823369
36. Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol. 2004;4:641–648. doi:10.1038/nri1415
37. Yin T, He S, Liu X, et al. Extravascular red blood cells and hemoglobin promote tumor growth and therapeutic resistance as endogenous danger signals. J Immunol. 2015;194:429. doi:10.4049/jimmunol.1400643
38. Carr BI, Cavallini A, D’Alessandro R, et al. Platelet extracts induce growth, migration and invasion in human hepatocellular carcinoma in vitro. BMC Cancer. 2014;14. doi:10.1186/1471-2407-14-43
39. Murciano-Goroff YR, Warner AB, Wolchok JD, et al. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30:507–519. doi:10.1038/s41422-020-0337-2
40. Bagchi S, Yuan R, Engleman EG, et al. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–249. doi:10.1146/annurev-pathol-042020-042741
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.