Back to Journals » International Journal of Nanomedicine » Volume 12
Cisplatin-resistant lung cancer cell–derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100–5p-dependent manner
Authors Qin X, Yu S, Zhou L, Shi M , Hu Y, Xu X, Shen B, Liu S, Yan D, Feng J
Received 3 January 2017
Accepted for publication 3 April 2017
Published 15 May 2017 Volume 2017:12 Pages 3721—3733
DOI https://doi.org/10.2147/IJN.S131516
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Xiaobing Qin,1,2,* Shaorong Yu,1,3,* Leilei Zhou,1,4 Meiqi Shi,3 Yong Hu,1 Xiaoyue Xu,1 Bo Shen,1 Siwen Liu,1 Dali Yan,1 Jifeng Feng1,3
1Research Center for Clinical Oncology, Affiliated Cancer Hospital of Nanjing Medical University, Jiangsu Cancer Hospital and Jiangsu Institute of Cancer Research, Nanjing, 2Department of Oncology, Xuzhou First People’s Hospital, Xuzhou, 3Department of Oncology, Affiliated Cancer Hospital of Nanjing Medical University, Jiangsu Cancer Hospital and Jiangsu Institute of Cancer Research, Nanjing, 4Department of Oncology, Affiliated Huai’an Hospital of Nanjing Medical University, Huai’an, Jiangsu, China
*These authors have contributed equally to this work
Abstract: Exosomes derived from lung cancer cells confer cisplatin (DDP) resistance to other cancer cells. However, the underlying mechanism is still unknown. A549 resistance to DDP (A549/DDP) was established. Microarray was used to analyze microRNA (miRNA) expression profiles of A549 cells, A549/DDP cells, A549 exosomes, and A549/DDP exosomes. There was a strong correlation of miRNA profiles between exosomes and their maternal cells. A total of 11 miRNAs were significantly upregulated both in A549/DDP cells compared with A549 cells and in exosomes derived from A549/DDP cells in contrast to exosomes from A549 cells. A total of 31 downregulated miRNAs were also observed. miR-100–5p was the most prominent decreased miRNA in DDP-resistant exosomes compared with the corresponding sensitive ones. Downregulated miR-100–5p was proved to be involved in DDP resistance in A549 cells, and mammalian target of rapamycin (mTOR) expression was reverse regulated by miR-100–5p. Exosomes confer recipient cells’ resistance to DDP in an exosomal miR-100–5p-dependent manner with mTOR as its potential target both in vitro and in vivo. Exosomes from DDP-resistant lung cancer cells A549 can alter other lung cancer cells’ sensitivity to DDP in exosomal miR-100–5p-dependent manner. Our study provides new insights into the molecular mechanism of DDP resistance in lung cancer.
Keywords: lung cancer, cisplatin, exosome, miR-100–5p, drug resistance
Introduction
Lung cancer is a serious threat to people’s lives and health worldwide.1 Surgical treatment, chemotherapy, radiotherapy, and target therapy are the most effective lung cancer treatment methods. Unfortunately, the outcome among lung cancer patients remains unsatisfactory. The most vital cause of treatment failure is the lung cancer cells’ resistance to treatment.
Exosomes are 30–100 nm nanosized vesicles and contain multiple materials, such as protein, DNA, messenger RNA (mRNA), microRNA (miRNA), and long noncoding RNA. They are widely distributed in various human body fluids, such as blood plasma/serum, saliva, breast milk, cerebrospinal fluid, and urine. The main function of exosomes is to participate in cell-to-cell communication.2 Recent literatures showed that exosomes are closely related to tumor development and metastasis and even influence the therapeutic effect in cancer patients.3–5
MiRNAs are a class of 17–24nt small, noncoding RNAs that mediate post-transcriptional gene silencing by binding to the 3′-untranslated region (3′-UTR) or the open reading frame region of target mRNAs. MiRNAs regulate a wide range of physiological and pathological processes, including cell proliferation, differentiation, and migration, as well as disease initiation and progression.6,7 The stable presence of miRNAs in body fluids is partly because they are packed into microvesicles or exosomes.8 Besides, miRNAs and mRNAs in exosomes are functional. They can be delivered to another cell and be functional in new location. For example, after transfer of mouse exosomal RNA to human mast cells, new mouse proteins can be found in the recipient cells.9 Therefore, the roles that miRNAs play in exosomes are gaining increased attention.10–12
Recently, accumulating evidence proved that exosomes could participate in communication of tumor cells.13,14 Corcoran et al found that exosomes may play an important role in docetaxel resistance of prostate cancer in vitro observations.15 In 2014, we reported that lung cancer cells A549 can secrete much more exosomes to the surrounding microenvironment after cisplatin (DDP) treatment, and these exosomes can reduce sensitivity of receptor cells to DDP.16 More importantly, exosomes-shuttling miRNAs probably conferred this resistance delivery. Followed by these studies, more and more evidence confirmed this observation in other cancers including breast cancer, prostate cancer, hepatocellular cancer,17–20 and so on. However, similar to our previous study, most of these literatures mainly reported this phenomenon and few researchers13 conduct in-depth study and systematically explore its detailed mechanism. In the present study, we carried out the in-depth research in lung cancer cells and proved that DDP-resistant lung cancer cell–derived exosomes contain miRNAs, which were characterized by prominent low expression of miR-100–5p and can alter recipient lung cancer cells’ resistance to DDP by increasing mammalian target of rapamycin (mTOR) expression of recipient cells.
Materials and methods
Cell culture
Human lung adenocarcinoma (LAC) cells A549 were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). A549 resistance to DDP (A549/DDP) was successfully established from A549 by exposing A549 to gradually increasing DDP concentrations, until the final concentration of DDP is 1 μg/mL. All cells were grown in Roswell Park Memorial Institute (RPMI)-1640 medium (Nanjing Kaiji Biology, Nanjing, China) containing 10% fetal bovine serum (FBS) (GIBCO, New York, NY, USA). Cells were incubated at 37°C and 5% CO2 in a humidified atmosphere. The cell lines were last authenticated by short tandem repeat DNA fingerprinting on October 18, 2014. To avoid the influence of DDP to A549/DDP cells, cells were cultured in a drug-free medium for at least 2 weeks before subsequent experiments.
Isolation of exosomes
Exosome-depleted FBS was used in the following experiments to avoid the impact of exosomes. FBS was depleted of exosomes by ultracentrifugation at 1×106 g at 4°C for 16 h (Beckman Coulter Avanti J-30I, USA). After being incubated for 48–72 h, the culture medium was harvested and exosomes were isolated by ultracentrifugation. Briefly, cell culture medium was sequentially centrifuged at 300 g for 10 min, 2,000 g for 15 min, and 12,000 g for 30 min to remove floating cells and cellular debris. These were then passed through a 0.22-μm filter. The supernatant was further ultracentrifuged at 1×106 g for 2 h at 4°C, washed in phosphate-buffered saline (PBS), and submitted to a second ultracentrifugation in the same conditions. Exosomes were quantified with bicinchoninic acid (BCA) method. Exosomal protein was measured by BCA protein assay kit (Beyotime Biotechnology, Nantong, China). The final exosome pellets were used immediately.
Transmission electron microscope
Exosomes were precipitated and immediately fixed in 2.5% glutaraldehyde at 4°C for the electron microscope observation. After fixation, specimens were processed through dehydration in gradient alcohol, and infiltrated in epoxy resin and then embedded. The ultrathin sections were stained with uranyl acetate and lead citrate, and were observed under transmission electron microscope (TEM) (JEM-1010; JEOL, Tokyo, Japan).
Western blot
Cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology). Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. Rabbit polyclonal CD63 (SBI, CA, USA) and mTOR antibody (Cell Signaling Technology, Boston, MA, USA) were used at a dilution of 1:1,000. β-actin (Cell Signaling Technology) was used at 1:1,000 dilution. The bound antibodies were detected using electrochemiluminescence (ECL) Western Blotting Detection system.
Co-culture assay
Exosomes were added separately into the fresh media of cells, at a dose of 100 μg. The same amount of PBS was placed into the media and the cells were cultured under the same conditions as control. After 48 h, each sample of cells was used for the next experiment.
Cell Counting Kit-8 assay
Cell Counting Kit-8 (CCK-8) assay was used to measure cells’ sensitivity to drug. Cells were seeded into 96-well plates at a density of 5×103 cells/well. Different DDP concentrations were added into culture media for 48 h. Ten microliters of freshly prepared CCK-8 solutions (Dojindo, Kumamoto, Japan) and 100 μL incomplete culture medium were added into each well. The optical density was measured at 450 nm using a scanning multiwell spectrophotometer (Bio-Rad Model 550; Bio-Rad, Hercules, CA, USA) after 1 hour. Cell growth inhibition curve was established, and the inhibitory concentration to produce 50% cell death (IC50) of DDP was calculated. All experiments were performed in triplicate.
Cell apoptosis and cell-cycle assays
The Annexin-V–fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA) was used to evaluate the apoptotic rate for the analysis according to the manufacturer’s instructions. The Annexin-V–FITC binding on cells was analyzed by flow cytometry (BD FACSCalibur, San Diego, CA, USA). All experiments were repeated triplicate.
Single-cell suspension of each sample was fixed at 1 mL 75% alcohol, and stored at −20°C overnight for the cell-cycle analysis. The G0/G1, S, and G2/M phase fractions were then determined by flow cytometry (BD FACSCalibur). All experiments were performed in triplicate.
RNA exaction from cells and exosomes for microarray analysis
As in our previous work,21 miRNA expression profiles of lung cancer cells A549 and its DDP-resistant cells A549/DDP were analyzed by microarray. MiRNA expression profiles of exosomes derived from A549 and A549/DDP were also analyzed by microarray. The microarray experiment was performed by the BGI Company (Beijing, China). Exosomal RNA was extracted by Total RNA Purification Kit (Norgen, Thorold, ON, Canada) according to the manufacturer’s protocols, and total RNA of cells was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). RNA was quantified by NanoDrop® ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA), and its quality was assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Human microRNA Microarray Kit (Agilent Technologies) was used for labeling and hybridization according to its instructions. The expression profiling of miRNA was conducted using the Agilent Human 8×60 K. The differential expression of miRNAs was screened to exclude those <1.5-fold changes in contrast to A549 or A-exo (exosomes of A549). The microarray data have been submitted to the Gene Expression Omnibus and the data could be accessed by the accession numbers GSE85603 and GSE85604.
Transfection experiment
Cells were seeded into six-well plates 1 day ahead of schedule and incubated until the 50% confluency was reached. miR-100–5p mimic, inhibitors, and negative control were purchased from Ribobio Company (Guangzhou, China). The miR-100–5p mimic, inhibitors, and negative control were mixed with 5 μL of Lipofectamine 2000 (Invitrogen), respectively, and then transfected into cells according to the Lipofectamine 2000 operating instructions. After 6 h of transfection, the liquid was abandoned and replaced with fresh complete medium. Cells were used for the next experiment after 48 h of transfection.
Real-time quantitative reverse transcriptase-polymerase chain reaction
The Total Exosome RNA and Protein Isolation Kit (Invitrogen) was used to extract exosomal RNA according to its instructions. Total RNA of cells was isolated using Trizol Reagent (Invitrogen). Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was determined by SYBR green technique. U6 snRNA was used as an endogenous control to normalize miRNA expression in cells, and cel-miR-39 was used to normalize miRNA expression in exosomes between the samples. β-actin was used as the endogenous control to normalize mTOR expression. The primers of miRNAs were synthesized by RiboBio Company, and the mTOR primers were synthesized by Sangon Biotech (Shanghai, China). Relative miRNA and mRNA expression levels were calculated by the 2−ΔΔCt method. All samples were tested thrice.
In vivo chemosensitivity assay
All animal experiments were undertaken in accordance with the Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of the affiliated cancer hospital of Nanjing Medical University. BABL/c athymic nude mice (male, 5 weeks old) were purchased from the Shanghai Experimental Animal Center of Chinese Academy of Sciences (Shanghai, China) and bred at pathogen-free conditions in the Animal Center of Nanjing Medical University. A total of 1×106 A549 cells were injected subcutaneously into the right flank of each mouse. Animals were randomly divided into five groups with five nude mice in each group. When tumors grew to 150–200 mm3, nude mice were administered with an intraperitoneal injection of DDP, 4 mg/kg, twice a week. At the same time, exosomes extracted with ultracentrifugation from the culture medium were directly injected into the implanted tumors at the dose of 100 μg per mouse every 3 days for 7 times. Tumor volume (V) was monitored by measuring length (L) and width (W) and calculated with the formula V = (L×W2)×1/2. Mice were killed 21 days after DDP injection. Tumors were separated, weighted, and measured, and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), Western blot, and qRT-PCR were used to detect the apoptosis rate, mTOR protein, and mRNA expression level, respectively.
Immunohistochemistry
Immunohistochemistry (IHC) was conducted to analyze mTOR protein expression. The percentage of positive tumor cells was calculated by image analysis using Image-Pro Plus 4.5 (Media Cybernetics, Silver Spring, MD, USA). Tissues were photographed using a Leica microscope (Leica Camera, Solms, Germany).
Statistical methods
All data were representative of at least three independent experiments and were presented as mean ± standard deviation. Student’s t-tests and one-way analysis of variance were used to analyze results. P<0.05 was considered statistically significant. All of the statistical analyses were performed with the Graphpad 5.0 and the SPSS 22.0 software.
Results
Characterization of isolated exosomes
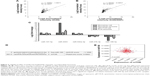
To ensure successful isolation of exosomes, the collected exosomes were observed by TEM and analyzed by Western blot. The vesicles appeared to be round and the diameter was between 30 and 100 nm under TEM (Figure 1A). Western blot showed that CD63 was expressed in exosomes, undetected in cells (Figure 1B).
Exosomes can confer drug resistance
IC50 of A549 and A549/DDP cells to DDP were 1.93±0.53 μg/mL and 21.15±1.07 μg/mL, respectively. In the following studies, the samples were divided into three groups: A549 + PBS, A549 + A-exo (A549 incubated with exosomes from A549), and A549 + R-exo (A549 incubated with exosomes from A549/DDP). A-exo (exosomes of A549) and R-exo (exosomes of A549/DDP) were added into the fresh media, respectively. The same amount of PBS was used as control and cultured under the same conditions. After 48 h, each sample was treated with DDP for 48 h. After incubating with exosomes, the CCK-8 assay results revealed that IC50 of A549 incubated with R-exo against DDP was 2.80±0.51 μg/mL, A549 incubated with A-exo was 2.10±0.39 μg/mL, and the control was 1.93±0.46 μg/mL. The IC50 of A549 incubated with R-exo was 33.3% higher than A549 with A-exo (P=0.03); however, the IC50 between A549 with or without A-exo were not significantly different (P>0.05; Figure 1C).
In the next apoptosis experiment, the results showed that the apoptotic rate of A549 with R-exo induced by DDP was reduced (38.0%) compared to A549 with A-exo (P=0.02), whereas the apoptotic rates of A549 with PBS were not significantly different from A549 with A-exo (P>0.05; Figure 1D). Incubation of A549 with R-exo increased G1 phase and decreased S phase relative to A549 with PBS, whereas the incubation with A-exo increased S phase in respect to A549 (P<0.05; Figure 1E). These results demonstrated that the exosomes from drug-resistant cells confer drug resistance to other cancer cells.
MiRNA profiles of A549 and A549/DDP, and their corresponding exosomes
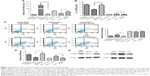
We used miRNA microarray to discover the difference expression of miRNAs between A549 cells and A549/DDP cells and their corresponding exosomes. As in our previous work,21 significant correlation between the miRNA profiles of A549 cells and their exosomes was detected by linear regression analysis (Figure 2A). The miRNA profiles of A549/DDP cells and exosomes showed similar results (Figure 2B). Intersection of miRNAs that are significantly upregulated both in A549/DDP cells compared with A549 cells and in R-exo in contrast to A-exo was analyzed. Intersection of significantly downregulated miRNAs was examined as well. Eleven upregulated miRNAs and 31 downregulated miRNAs (P<0.05) were detected eventually. Among the top 10 differentially expressed miRNAs (Table 1), miR-4443 was the most noticeable elevated miRNA, and miR-100–5p was the most prominent decreased miRNA in A549/DDP exosomes. Afterwards, we conducted qRT-PCR to examine the expression levels of the top five differential expressed miRNAs between lung cancer cells A549 and A549/DDP, and exosomes derived from A549 and A549/DDP, namely, miR-197–5p, miR-4443, miR-642a-3p, miR-27b-3p, and miR-100–5p. As shown in Figure 2C, results of qRT-PCR were in agreement with microarray. To further investigate the DDP resistance mechanisms, miR-100–5p was chosen for in-depth research.
  | Table 1 The top ten differentially expressed miRNAs in cells and exosomes |
mTOR may be a target gene of miR-100–5p in LAC
Several bioinformatics methods, including TargetScan, microRNA.org, and miRDB, were used to identify the potential target genes of miR-100–5p, and mTOR was selected. As shown in Figure 2D, the 3′-UTR of mTOR contains putative miR-100 complementary sites and the mirSVR score is −1.1698 and the PhastCons score is 0.6882. Besides, several studies22,23 illuminated that miR-100–5p can regulate the expression of mTOR by directly binding to the 3′-UTR of mTOR by the luciferase reporter.
To evaluate the relationship of mTOR and miR-100–5p in LAC cases, we collected all the miR-100–5p and mTOR expressions of LAC cases in The Cancer Genome Atlas (TCGA) database (n=527). A significant negative correlation between miR-100–5p and mTOR in LAC was observed (P=0.045; Figure 2E). These results further confirmed that mTOR acts as a target gene of miR-100–5p in LAC.
miR-100–5p alters DDP sensitivity of lung cancer cells
We first need to confirm whether absence of miR-100–5p is involved in DDP resistance in lung cancer cells. We confirmed that the miR-100–5p expression level in A549/DDP was lower than that in A549 by qRT-PCR. The expression level increased dramatically (P<0.001) once transfected with miR-100–5p mimic in A549/DDP. On the contrary, after being transfected with miR-100–5p inhibitor in A549, the expression level significantly reduced (P=0.012; Figure 3A). The IC50 of A549/DDP cells transfected with miR-100–5p mimics was lower than that in the negative control (P=0.014). The IC50 of A549 transfected with miR-100–5p inhibitors was higher than that in the negative control (P=0.03; Figure 3B).
The apoptosis of A549/DDP cells transfected with miR-100–5p mimics was obviously greater than the negative control (P=0.003). A549 transfected with miR-100–5p inhibitors has an apoptotic rate significantly less than that in the negative control (P=0.03; Figure 3C). These results indicate that miR-100–5p was essential for A549 cells’ response to DDP. Furthermore, Figure 3D and E showed that the protein expressions and the mRNA levels of mTOR were downregulated in A549/DDP cells transfected with miR-100–5p mimics compared with the negative control, and increased in A549 cells transfected with miR-100–5p inhibitors compared with the negative control. These results indicated that miR-100–5p expression could change A549 cells’ resistance to DDP through mTOR.
MiR-100–5p in exosomes influence DDP resistance of recipient cells
We screened exosomal miRNA expression of cells by qRT-PCR. Results indicated that miR-100–5p was downregulated by 75% in A549/DDP exosomes than that in A549 exosomes. As shown in Figure 4A, the relative miR-100–5p expression level of exosomes from A549/DDP transfected with miR-100–5p mimic was significantly higher than that transfected with miR-100–5p control mimic (P<0.001), and the fold change is larger than that in cells. The relative miR-100–5p expression level of exosomes from A549 transfected with miR-100–5p inhibitor was lower than that transfected with miR-100–5p control inhibitor (P=0.008). However, no significant difference in the expression level of exosomal mTOR between the groups was observed (not shown). Thus, these results prove that miR-100–5p can be packaged into exosomes.
In the following research, six groups were prepared as below: group a, A549 incubated with A549/DDP exosomes; group b, A549 incubated with exosomes of A549/DDP transfected with miR-100–5p mimic; group c, A549 incubated with exosomes of A549/DDP transfected with miR-100–5p control mimic; group d, A549 incubated with A549 exosomes; group e, A549 incubated with exosomes of A549 transfected with miR-100–5p inhibitor; group f, A549 incubated with exosomes of A549 transfected with miR-100–5p control inhibitor. As determined by qRT-PCR, the miR-100–5p expression level in group b was increased remarkably than group c (P<0.001), and the expression level in group e was decreased dramatically than group f (P=0.015; Figure 4B). The apoptosis rate of group b was evidently greater than that of group c, and the apoptosis rate of group e was less than that of group f (Figure 4C). The IC50 of group b was lower than group c, and the IC50 of group e was higher than group f (P<0.05; Figure 4D). Moreover, the mRNA and protein levels of mTOR were decreased in group b in contrast to group c, and were increased in group e compared with group f (P<0.05; Figure 4E and F). These results indicated that DDP resistance can be transmitted in an exosomal miR-100–5p-dependent manner, at least partly by modulating the expression of mTOR.
Exosomal miR-100–5p alters DDP resistance in vivo
After inoculation, the transplanted nude mice were randomly divided into five groups, and the following are the respective intratumoral injection reagents: group 1: PBS; group 2: (A549/DDP + miR-100–5p mimic)-exo (exosomes of A549/DDP transfected with miR-100–5p mimic); group 3: (A549/DDP + miR-100–5p control mimic)-exo (exosomes of A549/DDP transfected with miR-100–5p control mimic); group 4: (A549 + miR-100–5p inhibitor)-exo (exosomes of A549 transfected with miR-100–5p inhibitor); group 5: (A549 + miR-100–5p control inhibitor)-exo (exosomes of A549 transfected with miR-100–5p control inhibitor). All nude mice were treated with DDP. It shows that the tumors of group 2 grew significantly slower than group 3 after the treatment with DDP, and the tumors of group 4 grew remarkably faster than group 5 (Figure 5A). At 35 days after inoculation, the tumor volume and weight of group 3 were significantly larger than group 2 following the DDP treatment (P<0.05), and downregulated exosomal miR-100–5p led to the resistance of cancer cells to DDP, and then promoted the tumor growth (P<0.05) (Figure 5B and C). We subsequently found that in group 2 the miR-100–5p expression was significantly upregulated and mTOR expression significantly decreased than the negative control (P<0.01; Figure 5D and E). The IHC showed that the mTOR protein positivity in group 2 tumor was significantly weaker than that in group 3 (Figure 5F). TUNEL assay showed that, after DDP treatment, the apoptotic rate of xenografts developed from group 2 was remarkably greater than group 3 (P<0.01), and group 4 was significantly lower than their negative control group (P<0.05; Figure 5G).
Discussion
In the present study, we proved that DDP-resistant A549 cell–derived exosomes, which were characterized by prominent low expression of miR-100–5p, confer other cancer cells’ resistance to DDP in an exosomal miR-100–5p-dependent manner both in vitro and in vivo. This was mainly achieved by modulating mTOR expression of recipient cells.
Recently, there has been a growing number of evidence showing that exosomes can participate in drug resistance.15,24 Our results, along with the previous study, suggest that exosomes from DDP-resistant lung cancer cells (A549/DDP) confer recipient cells’ resistance to DDP. Moreover, according to microarray results, miR-100–5p was the most prominent decreased miRNA in DDP-resistant exosomes, and the fold change of miR-100–5p in exosomes was larger than that in cells, which implied that this abnormal reverse concentration of miR-100–5p might play a vital role in communication between tumor cells. More importantly, the decreased expression of miR-100 was wildly reported to be involved in drug resistance in various cancer patients (we will discuss this below). Therefore, we chose miR-100–5p for an in-depth research.
MicroRNA-100 (miR-100), located on chromosome 11 at 11q24.1, is a member of the miR-99 family. It is involved in regulating multiple cell processes, such as cell apoptosis, cell cycle, differentiation, proliferation, invasion, and migration.25,26 miR-100 was reported to enhance the chemosensitivity of various tumor cells to chemotherapeutics.27,28 Several researches proved that overexpression of miR-100 can sensitize cancer cells to DDP in various tumors.23,29 Concurring with these studies, our research confirmed that the downregulated expression of miR-100–5p contributed to DDP resistance in lung cancer cells. The luciferase reporter in previous literatures22,23 proved that mTOR is a direct target gene of miR-100–5p and can be regulated by directly binding to the 3′-UTR of mTOR. In our study, we did find that mTOR expression was negatively regulated by miR-100–5p, thereby modulating the sensitivity of lung cancer cells to DDP. Therefore, we believed that mTOR is at least one of the target genes of miR-100–5p and can be regulated by miR-100–5p and then affects the sensitivity of lung cancer cells to DDP.
It is naturally accepted that lower expression of miR-100–5p in cells can result in higher expression of mTOR and consequently lead to cancer cells’ resistance to DDP. The question is how exosomes containing low-expression miR-100–5p can affect other cells’ resistance to DDP. A possible explanation was that tumor cells can be regulated by miRNA both in themselves and their surrounding environment. In other words, lung cancer cells’ sensitivity to DDP can be regulated by miR-100–5p not only in cells but also outside of cells. It seems that miR-100–5p outside can support miR-100–5p inside to display its function well. If the surrounding environment is enriched with miR-100–5p, these miRNAs could be absorbed by recipient cells and together with miRNA inside to display their function. However, once the surrounding environment lacks “normal” miR-100–5p existence, the function of miR-100–5p inside discounts greatly. In fact, our in vivo study found that miR-100–5p expression in tumor cells was affected by miR-100–5p in exosomes: when there are abundant miR-100–5p in exosomes, the miR-100–5p in cells was extremely high, whereas when miR-100–5p in exosomes was inhibited, and the miR-100–5p in cells was slightly decreased.
Target therapies have been developed in recent years; however, survival rates are not improving because of the delay in the diagnosis, which makes biomarkers one of the most interesting fields of study in cancer. DDP is one of the main drugs used in clinic. There are plenty of studies to identify DDP responders from the whole lung cancer patients and predict the treatment outcome of patients.30–32 However, most of them failed to demonstrate the predictive role in phase III clinical trial.33 The reason why these biomarkers could not get a positive result in clinical trial was probably because these biomarkers could only reflect the static state of tumor cells before chemotherapy. In fact, tumor cells were not static; they need to change to adapt to their surroundings. In this circumstance, we need a dynamic biomarker to predict patients’ response to DDP and exosome is an optimal alternative. Consistent with other studies,9,34 we proved that exosome components come from original cells and at least partly reflect the content of the original tumor cells, which indicates that exosomes might be a perfect candidate to predict DDP chemosensitivity in lung cancer patients in the future.35
In summary, our study proved that DDP-resistant lung cancer cell–derived exosomes characterized by low expression of miR-100–5p confer recipient cells’ resistance to DDP in an exosomal-miR-100–5p-dependent manner. Exosomal miR-100–5p can affect expression of mTOR in recipient cells and thereby modulate cells’ sensitivity to DDP. As miRNA expression in exosomes can partly represent miRNA expression of maternal cells, exosomes derived from tumor cells provide a potential new dynamic biomarker to tailor chemotherapy of lung cancer patients in the future.
Acknowledgments
We are grateful for financial supports from the National Natural Science Foundation of China (Nos 81402483 and 81372396); the Project of Invigorating Health Care through Science, Technology and Education, Jiangsu Provincial Medical Youth Talent (No QNRC2016646); Natural Science Foundation of Jiangsu province (No bk20141017); Xuzhou science and technology plan project of China (No KC16SH030); Jiangsu University Clinical Medical Science and Technology Development Fund of China (No JLY20160122). We thank Jianzhong Wu and Haixia Cao for providing us with general support and technical help.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6(35):37151–37168. | ||
Li L, Li C, Wang S, et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76(7):1770–1780. | ||
Felicetti F, Feo AD, Coscia C, et al. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J Transl Med. 2016;14:56. | ||
Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol Sci. 2016;37(7):606–617. | ||
Pastorkova Z, Skarda J, Andel J. The role of microRNA in metastatic processes of non-small cell lung carcinoma. A review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160(3):343–357. | ||
Larrea E, Sole C, Manterola L, et al. New concepts in cancer biomarkers: circulating miRNAs in liquid biopsies. Int J Mol Sci. 2016;17(5):1–42. | ||
Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184(1):28–41. | ||
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. | ||
Yuan D, Xu J, Wang J, et al. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget. 2016;7(22):32707–32722. | ||
Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139(7):1443–1448. | ||
Liao J, Liu R, Shi YJ, Yin LH, Pu YP. Exosome-shuttling microRNA-21 promotes cell migration and invasion-targeting PDCD4 in esophageal cancer. Int J Oncol. 2016;48(6):2567–2579. | ||
Le Q, Jin D, Cheng C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. | ||
Qu Z, Wu J, Luo D, Jiang C, Ding Y. Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J Exp Clin Cancer Res. 2016;35(1):159. | ||
Corcoran C, Rani S, O’Brien K, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7(12):e50999. | ||
Xiao X, Yu S, Li S, et al. Exosomes: decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One. 2014;9(2):e89534. | ||
Kong JN, He Q, Wang G, et al. Guggulsterone and bexarotene induce secretion of exosome-associated breast cancer resistance protein and reduce doxorubicin resistance in MDA-MB-231 cells. Int J Cancer. 2015;137(7):1610–1620. | ||
Mao L, Li J, Chen WX, et al. Exosomes decrease sensitivity of breast cancer cells to adriamycin by delivering microRNAs. Tumour Biol. 2016;37(4):5247–5256. | ||
Corcoran C, Rani S, O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014;74(13):1320–1334. | ||
Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. | ||
Qin X, Yu S, Xu X, Shen B, Feng J. Comparative analysis of microRNA expression profiles between A549, A549/DDP and their respective exosomes. Oncotarget. Epub 2017 Feb 2. | ||
Xu C, Zeng Q, Xu W, et al. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther. 2013;12(2):207–219. | ||
Zhu Z, Wang CP, Zhang YF, Nie L. MicroRNA-100 resensitizes resistant chondrosarcoma cells to cisplatin through direct targeting of mTOR. Asian Pac J Cancer Prev. 2014;15(2):917–923. | ||
Lv MM, Zhu XY, Chen WX, et al. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumour Biol. 2014;35(11):10773–10779. | ||
Li C, Gao Y, Zhang K, et al. Multiple roles of microRNA-100 in human cancer and its therapeutic potential. Cell Physiol Biochem. 2015; 37(6):2143–2159. | ||
Luan Y, Zhang S, Zuo L, Zhou L. Overexpression of miR-100 inhibits cell proliferation, migration, and chemosensitivity in human glioblastoma through FGFR3. Onco Targets Ther. 2015;8:3391–3400. | ||
Zhang B, Zhao R, He Y, et al. MicroRNA 100 sensitizes luminal A breast cancer cells to paclitaxel treatment in part by targeting mTOR. Oncotarget. 2016;7(5):5702–5714. | ||
Luo J, Chen B, Ji XX, Zhou SW, Zheng D. Overexpression of miR-100 inhibits cancer growth, migration, and chemosensitivity in human NSCLC cells through fibroblast growth factor receptor 3. Tumor Biol. 2015;25:252–254. | ||
Liu Y, Zhu ST, Wang X, et al. MiR-100 inhibits osteosarcoma cell proliferation, migration, and invasion and enhances chemosensitivity by targeting IGFIR. Technol Cancer Res Treat. 2016;15(5):NP40–NP48. | ||
Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25(19):2747–2754. | ||
Bonanno L, Costa C, Majem M, Favaretto A, Rugge M, Rosell R. The predictive value of BRCA1 and RAP80 mRNA expression in advanced non-small-cell lung cancer patients treated with platinum-based chemotherapy. Ann Oncol. 2013;24(4):1130–1132. | ||
Wang B, Matsuoka S, Ballif BA, Zhang D, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–1198. | ||
Moran T, Wei J, Cobo M, et al. Two biomarker-directed randomized trials in European and Chinese patients with nonsmall-cell lung cancer: the BRCA1-RAP80 Expression Customization (BREC) studies. Ann Oncol. 2014;25(11):2147–2155. | ||
Xiao D, Ohlendorf J, Chen Y, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One. 2012;7(10):e46874. | ||
Reclusa P, Sirera R, Araujo A, et al. Exosomes genetic cargo in lung cancer: a truly Pandora’s box. Transl Lung Cancer Res. 2016;5(5):483–491. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.