Back to Journals » OncoTargets and Therapy » Volume 13
Circulating Tumor Cells as a Screening and Diagnostic Marker for Early-Stage Non-Small Cell Lung Cancer
Authors Duan GC , Zhang XP, Wang HE, Wang ZK, Zhang H, Yu L, Xue WF, Xin ZF, Hu ZH, Zhao QT
Received 11 December 2019
Accepted for publication 26 February 2020
Published 4 March 2020 Volume 2020:13 Pages 1931—1939
DOI https://doi.org/10.2147/OTT.S241956
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Guo-Chen Duan, Xiao-Peng Zhang, Hui-En Wang, Zhi-Kang Wang, Hua Zhang, Lei Yu, Wen-Fei Xue, Zhi-Fei Xin, Zhong-Hui Hu, Qing-Tao Zhao
Department of Thoracic Surgery, Hebei General Hospital, Shijiazhuang, Hebei, People’s Republic of China
Correspondence: Qing-Tao Zhao
Department of Thoracic Surgery, Hebei General Hospital, 348, West He-Ping Road, Shijiazhuang 050051, Hebei Province, People’s Republic of China
Tel +86-31185988756
Email [email protected]
Background: Circulating tumor cells (CTCs) have become potential diagnostic biomarker for several types of cancer, including lung cancer. In this study, we aim to determine whether CTCs detected by CellCollector can be used for early-stage diagnosis of lung cancer.
Methods: In this study, we recruited 64 volunteers, among whom 44 were suspected lung cancer patients requiring surgical treatment and 20 were healthy volunteers. We simultaneously analyzed PD-L1 expression in CTCs isolated using the GILUPI CellCollector and copy number variation by next-generation sequencing (NGS).
Results: We enrolled a total of 44 patients with suspected lung cancer who required surgery and 20 healthy volunteers. The patients were classified into 4 groups based on their pathological results: benign disease, in situ cancer, microinvasive, and invasive. The CTCs detection rate for each group was 10.00% (1/10), 45% (5/11), 50% (7/14), and 67% (6/9), respectively. Among the patients with lung cancer, the CTCs detection rate increased with disease progression. The rate of CTCs positivity was 52.94% (18/34) in patients who were diagnosed with lung cancer by pathology and 10% (1/10) in patients with benign disease. CTCs were not detected in the control group. The area under the receiver operating characteristic (ROC) curve, a measure for distinguishing patients with primary lung cancer, was 0.715 (95% CI 0.549– 0.880, P=0.041). The sensitivity and specificity of the in vivo CTCs detection strategy for the diagnosis of early-stage lung cancer were 52.94% and 90%, respectively. CTCs were associated with clinical pathology but not with the size and location of the nodules.
Conclusion: CTCs isolation using the CellCollector in vivo detection method might be effective for distinguishing between benign and malignant nodules and may be used for early-stage diagnosis of lung cancer.
Keywords: CellCollector, in vivo, circulating tumor cells, early-stage diagnosis, lung cancer
Introduction
Lung cancer remains the most common malignant tumor worldwide. It is estimated that in 2018, 2.1 million patients will be newly diagnosed with lung cancer, and 1.8 million lung cancer patients will die. New lung cancer cases account for 11.6% of total cancer cases but 18.4% of total cancer deaths.1 Because the symptoms of lung cancer are not apparent in the early stages, the majority of lung cancer patients are at an advanced stage at the time of diagnosis and miss the opportunity for optimal treatment. As a result, the prognosis of lung cancer is extremely poor. The 5-year survival rate at stage IV is only 6%, yet the 5-year survival rate of patients with stage IA lung cancer is 82%.2 Therefore, the diagnosis of lung cancer at an early stage is a key factor for improving prognosis and reducing mortality. Low-dose spiral computed tomography (LDCT) scanning is a test that may increase early diagnosis and reduce cancer mortality. The National Lung Screening Trial (NLST) in the USA indicated that compared with thoracic X-ray imaging, LDCT reduces lung cancer-related mortality by 20%. However, LDCT screening has a high false-positive rate, and more than 10% of patients are overdiagnosed.3,4 Therefore, biomarkers with higher specificity and lower invasiveness are urgently needed for the early diagnosis of lung cancer.
Circulating tumor cells (CTCs) are a new type of tumor biomarker that offer the promise of real-time detection and minimal invasiveness. The examination of CTCs can provide powerful evidence and support for tumor evaluation, monitoring, treatment, and prevention. CTCs are tumor cells originating from tumor tissue that enter the peripheral blood circulation and are involved in tumor metastasis. CTCs are scarce in blood but have high clinical detection value and are an important marker for liquid biopsy.5 Currently, the techniques for capturing and isolating CTCs are based on their physical and chemical properties, such as cell size and density, or the expression of tumor cell surface proteins such as EpCAM for positive or negative enrichment.6 CTCs are normally examined in vitro, and the sensitivity of detection is often limited by the blood sample volume. The GILUPI CellCollector in vivo examination technique overcomes the limitation of a small blood sample volume and has a high detection rate at the early stage of cancer. This high detection rate makes this technique very favorable for application in the diagnosis of early-stage lung cancer.
It is generally believed that CTCs are produced in the early stages of tumors and represent tumor micrometastasis or the presence of small residual lesions. We hypothesized that a small number of CTCs are present in the peripheral blood of patients with confirmed lung cancer but may not in patients with benign disease. To test this hypothesis, this study recruited a total of 64 volunteers, including 44 suspected lung cancer patients requiring surgical resection and 20 healthy volunteers as a control. This study is the first to analyze PD-L1 expression in CTCs with the GILUPI CellCollector and the molecular characteristics of CTCs by next-generation sequencing (NGS). The sensitivity and specificity of the CTCs acquired by the CellCollector in the diagnosis of early-stage lung cancer were analyzed by postoperative pathological diagnosis.
Materials and Methods
Study Design
This was a single-center, prospective clinical study carried out in Hebei General Hospital, China. A total of 44 patients with suspected lung cancer were recruited between January 2018 and June 2018. All patients underwent surgical treatment and pathological diagnosis. Peripheral CTCs were detected using the CellCollector (GILUPI CellCollector, GILUPI) in vivo detection method prior to surgery. Immunofluorescence staining was performed for CTC evaluation and PD-L1 expression analysis in CTCs. Additionally, CTCs captured by the CellCollector sampling probe were separated and subjected to whole-genome amplification and quality assessment. Qualified CTCs underwent NGS to determine the copy number variation (CNV). Twenty healthy subjects were also recruited simultaneously for CTCs examination, and the basic information for these subjects, such as gender, age, and smoking status, was similar to that of the suspected lung cancer patients. This study was approved by the medical ethics committee of Hebei General Hospital, and all subjects signed informed consent. This study was conducted in accordance with the Declaration of Helsinki.
Patients
Among the 44 recruited patients with suspected lung cancer, patients diagnosed with solid nodules or partial solid nodules in the lung by thoracic computed tomography (CT) underwent positron emission tomography-computed tomography (PET-CT), or bone emission computed tomography (ECT), cranial magnetic resonance imaging (MRI) and abdominal CT examinations, and patients with ground-glass nodules in the lung did not undergo relevant imaging examinations. None of the patients had distant metastasis. Among the suspected lung cancer patients, 22 were male, and 22 were female; the median age was 56 years with an age range of 32 to 77 years, and the smoking rate was 25.00%. Among all patients, 5 had nodules at 2 or more locations, 13 had nodules in the upper lobe of the left lung, 10 had nodules in the left lower lobe, 12 had nodules in the right upper lobe, 8 had nodules in the right lower lobe, and 5 had nodules in the right middle lobe (Table 1). All these patients were pathologically diagnosed with non-small cell lung cancer (NSCLC), and all were adenocarcinoma (Table S1). The suspected lung cancer patients and healthy controls did not differ significantly with respect to age, gender, and history of smoking.
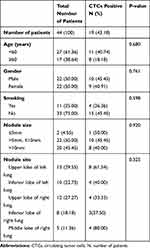 |
Table 1 Characteristics of Patients are Enrolled |
In vivo CTCs Detection by CellCollector
Cubital vein peripheral blood CTCs were examined by the circulating epithelial cell sampling probe of the CellCollector, a medical stainless steel wire with a 2-cm-long functional domain coated with EpCAM antibody and a hydrogel stratum. The sampling probe was inserted into cubital vein peripheral blood through a 20G catheter with the functional domain exposed to the peripheral blood and placed in vivo for 30 min. Tumor cells were captured by specific binding to the EpCAM antigen of the tumor cell surface (Figure 2A).
CTCs Analysis
Upon completion of CTCs collection, the CellCollector with captured CTCs underwent staining according to the instruction manual of the staining kit. Negative (NK92 cells) and positive (SK-BR-3 cells) controls were stained simultaneously. The following antibodies were used for staining: CD45 (EXBIO, clone MEM-28-Alexa Fluor 647), cytokeratin CK7/19/panCK antibody (EXBIO Praha, clone A53-B/A2-Alexa Fluor 488), and PD-L1 (clone PD-L1, Abcam). Following nuclear staining with Hoechst 33342 (Sigma), the cells were examined for tumor characteristics, and PD-L1 expression in CTCs was analyzed.
Whole-Genome Amplification and Next-Generation Sequencing
The CTCs captured in the CellCollector were identified and collected in PCR tubes. The isolated CTCs were then subjected to whole-genome amplification (WGA) using the MALBAC genome amplification kit (Yikon Genomics). The quality of the amplification products was determined using a NanoDrop 2000 and Qubit 3.0. The coverage of the amplification products was determined by fluorescence-based quantitative PCR. a) Amplification yield was qualified if the DNA concentration of single-cell amplification was ≥10 ng/μL and the total amount was ≥400 ng. b) A library could be constructed in the case of relatively low yield, with DNA concentrations in single-cell amplification of 1–10 ng/μL and total DNA amounts of 40–400 ng. c) Amplification failed if the DNA concentration of single-cell amplification was <1 ng/μL and the total amount was <40 ng. The qualified CTC genome amplification products were subjected to downstream gene mutation analysis. NGS was performed using the Illumina platform.
Statistical Analysis
Differences between the patients and healthy controls and the correlations between CTCs and clinical characteristics were determined using Student’s t test, Pearson’s Chi-square test, or Fisher’s exact test. P<0.05 was considered statistically significant. All statistical analyses were performed with SPSS 23.0 software. GraphPad Prism 5.0 software (GraphPad, San Diego, CA, USA) was used to generate the graphs.
Results
Subject Characteristics
A total of 44 suspected lung cancer patients and 20 healthy volunteers were recruited for CTCs examination. CTCs were not detected in the 20 healthy individuals of the control group. Among the 44 patients with suspected lung cancer who underwent surgical treatment, 34 patients were pathologically diagnosed with lung cancer, and the CTCs detection rate was 52.94% (18/34). The remaining 10 patients were pathologically diagnosed with benign lung diseases, of whom only 1 patient had CTCs (Figure 1).
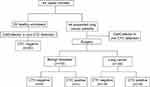 |
Figure 1 Experimental flow chart. |
CTCs Assessment by Staining
After capture by the CellCollector, the CTCs were examined by staining. White cells and tumor cells were determined using CD45 and CK7/19/panCK, respectively. CD45-, CK7/19/panCK+, Hoechst+ cells with an intact morphology were considered CTCs, and CD45+, CK7/19/panCK-, Hoechst+ cells were considered to be white cells. In the CTCs PD-L1 analysis, PD-L1+ indicated that PD-L1 was expressed in CTCs, and PD-L1- indicated that PD-L1 was not expressed in CTCs (Figure 2B). Figure 2C shows representative images of patients with positive CTCs (≥1 CTC) and the CTCs PD-L1 analysis. Among these patients, 2 CTCs were captured in patient 36, and PD-L1 was expressed in both CTCs. Three CTCs without PD-L1 expression were captured in patient 39. One CTC each was captured in patients 42 and 44, and PD-L1 was expressed in the CTCs of patient 44. Figure 2D shows the imaging and pathology results for patient 39. A thoracic CT scan identified a subpleural nodule in the right middle lobe and multiple micronodules in the right middle lobe, the posterior segment of the right upper lobe, and the lateral basal segment of the left lower lobe in this patient. The postoperative pathology showed that the patient had invasive adenocarcinoma. Elastic fiber staining showed that 70% of the adenocarcinoma was acinar, 20% was lepidic and 10% was papillary. Cancer cells were detected in the surrounding acinar cavity, and no clear cancer embolus and nerve invasion were observed. No pleural invasion was found.
Correlations Between CTCs and Clinical Features
To determine the correlations between CTCs and clinical features, we performed CTCs detection and analyzed the CTCs detection rate and CTCs counts. Our results showed that a total of 43.18% (19/44) patients were CTCs positive (≥1 CTC) and that the CTCs PD-L1 positive rate was 57.89% (11/19). No CTCs were detected in healthy controls (Figure 3A). Based on the pathology results, the patients were divided into the benign disease group and lung cancer group, for which the CTCs detection rates were 10.00% (1/10) and 52.94% (18/34), respectively (P=0.016) (Table 2 and Figure 3B). Based on lung cancer progression, the patients were divided into three groups, the in situ cancer group, microinvasive group, and invasive group, with CTCs detection rates of 45% (5/11), 50% (7/14), and 67% (6/9), respectively. Our results showed that in lung cancer patients, the CTCs detection rate and counts increased together with disease progression (Figure 3C). The area under the ROC curve distinguishing patients with primary lung cancer based on CTCs counts was 0.715 (95% CI 0.549–0.880, P=0.041) (Figure 3D). Based on these results, we found that the sensitivity and specificity of the in vivo CTCs detection strategy in early lung cancer diagnosis were 52.94% and 90%, respectively.
 |
Table 2 CTCs Detection in Different Pathology Group |
We further determined the correlation between patients’ characteristics and CTCs (Table 1). We found that the presence of CTCs was not correlated with gender, age, smoking history, nodule size, or the location of the patient. The rate of CTCs positivity was 50.00% (1/2) in the group with a nodule size ≤5 mm, 45.45% (10/22) in the group with a nodule size greater than 5 mm but no larger than 10 mm, and 40.00% (8/20) in patients with a nodule size >10 mm. The P value determined using the Chi-square test was 0.920. When the patients were divided into groups with different nodule locations, the rates of CTCs positivity for patients with nodules in the left upper lobe, left lower lobe, right upper lobe, right lower lobe, and right middle lobe were 61.54% (8/13), 40.00% (4/10), 33/33% (4/12), 37.50% (3/8), and 80.00% (4/5), respectively (P=0.325).
In the present study, we found that CTCs were associated with the clinical pathology but not with the nodule size or location. The rates of CTCs positivity were relatively higher in the left upper lobe and right middle lobe. However, only 5 of the 44 patients had nodules in the right middle lobe, and thus more data are required to confirm this result.
Whole-Genome Amplification and CNV Analysis of CTCs Captured in the CellCollector
The samples were subjected to single-cell whole-genome degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR) amplification and purification. Of the product, 1 μL was used for the Qubit test, and 5 μL was used for the caliper test. All samples were qualified (Table S2 and Figure S1). CNV and the corresponding changes in gene expression pattern were associated with malignant tumor occurrence and progression. To determine whether the presence of CTCs was associated with cancer, CNV status was analyzed by NGS in 4 randomly selected CTCs-positive patients (patient 21, patient 35, patient 41, patient 43) who were diagnosed with lung cancer by pathology and the only CTCs-positive patient with benign disease (patient 10). The sequencing results showed that all 5 samples had CNV (Figure S2). Analysis of the CNV and the corresponding mutated genes revealed some cancer-associated gene mutations, including SMAD4, MUTYH, SYK, and ARAMTS5.
Discussion
In recent years, some biomarkers have been identified in studies of early-stage lung cancer, including serum tumor markers, DNA methylation, circulating tumor DNA, and CTCs. Tumor markers can be used for the diagnosis of lung cancer patients and can be detected in the serum or plasma of patients with suspected tumors. Examination results may indicate the presence of tumors and predict tumor progression. For example, CYFRA21-1, CEA, and SCC are usually used for NSCLC, and NSE and ProGRP are usually used for small cell lung cancer (SCLC). However, due to the low sensitivity, specificity, and reproducibility of the identification of lung cancer serum biomarkers, it is still very difficult to use a single marker for cancer identification.7,8 Recently, genes with abnormal methylation have been found in lung cancer, and in the early stage of lung cancer, many cancer-related genes undergo methylation to varying degrees.9,10 However, it remains problematic to apply DNA methylation in lung cancer diagnosis due to its associated low positive rates for single genes and extremely limited highly specific sites. Other studies have indicated that the concentration of circulating tumor DNA (ctDNA) in cancer patients is significantly higher than that in the healthy population and that ctDNA can be used as a liquid biopsy for cancer diagnosis and prognosis. However, ctDNA levels in early-stage cancer are low, accounting for only 0.01–1% of cell-free DNA (cfDNA), and the presence of multiple techniques in the clinic and the lack of standardized methods limit its clinical application.11,12 Additionally, although CTCs have been reported in early cancer screening, further investigation is required to determine whether they can be used for early lung cancer diagnosis.
The GILUPI CellCollector is the only Class-III medical device approved by the National Medical Products Administration (NMPA) for clinical application in vivo. The CellCollector has high capture rates in multiple cancers, including lung cancer, breast cancer, prostate cancer, head and neck cancer, and neuroendocrine tumors, and its rate of CTCs positivity is approximately 70–80%.13–17 Moreover, using CellCollector, the detection rate in patients in early cancer stages is more than 50%, 20% higher than the detection rate of the similar product CellSearch.16,18,19 The high detection rate in early-stage tumors maybe facilitates research on the application of CTCs in the diagnosis of early-stage cancer. A study by He et al reported CTCs detection rate of 15.6% (5/32) in patients with ground-glass nodules and mutations in cancer-related genes, including KIT, SMARCBI, and AP53, in all CTCs-positive patients. However, that study did not include the pathological diagnosis results. LDCT combined with CTCs analysis has been shown to be useful for screening early-stage lung cancer, and the captured CTCs can be further used for downstream gene analysis.20
This study is the first to utilize the CellCollector in vivo detection system to assess CTCs in the diagnosis of early-stage lung cancer. In this study, we recruited 44 patients with lung nodules undergoing surgical resection and 20 healthy volunteers. Among the patients, 10 were diagnosed with benign disease and 34 with lung cancer based on postoperative pathology. The CTCs detection rates for the benign disease group and lung cancer group were 10.00% (1/10) and 52.94% (18/34), respectively, and no CTCs were detected in healthy volunteers. The sensitivity and specificity of the CellCollector in vivo CTCs detection technique in early-stage lung cancer diagnosis were 52.94% and 90%, respectively. The area under the ROC curve distinguishing patients with benign or malignant nodules using CTCs counts was 0.715 (P=0.041). The ROC curve indicated that the CTCs captured in vivo maybe used for the effective diagnosis of early-stage lung cancer. However, a previous study using the CellSearch system for early-stage lung cancer diagnosis indicated that CTCs captured in vitro could not effectively distinguish benign and malignant nodules, and the area under the ROC curve was 0.598 (P=0.122).21 This result might have been a consequence of the low sensitivity of the in vitro detection technique.
All patients were diagnosed with NSCLC based on postoperative pathology. According to the invasiveness of the nodules, we divided the lung cancer patients into the in situ cancer group, microinvasive group, and invasive group, with CTCs detection rates of 45.45%, 50.00% and 66.67%, respectively. The CTCs detection rate increased with the invasiveness of the nodules. In this study, we found that the CTCs detection rates in the left upper lobe and right middle lobe were relatively high, 61.54% and 80.00%, respectively. However, due to the limited patient population, we only recruited 5 patients with nodules in the right middle lobe, and the results did not reveal a correlation between nodule location and CTCs.
The PD-1/PD-L1 signaling pathway is involved in tumor immune escape. A study has shown that CTCs PD-L1-positive cells are more prone to immune escape and metastasis.22 Therefore, the inclusion of drug treatment and prognosis in the study of CTCs PD-L1 is of important clinical significance. However, although researchers have focused great attention on the detection of PD-L1 expression, reports on CTCs PD-L1 are very limited, mainly because CTCs are scarce and difficult to capture. Many techniques are not suitable for PD-L1 detection. The detection of CTCs by the CellCollector in vivo technique overcomes the difficulty of a limited sample size and allows for downstream protein analysis. In the current study, we detected CTCs PD-L1 expression, and the rate of CTCs PD-L1 positivity was 57.89% (11/19). Some studies have reported that NSCLC patients with positive CTCs PD-L1 have a poor prognosis.23,24 After a short-term follow-up of 17 months, all patients with benign diseases and lung cancer survived without progression. Follow-up data are required to confirm the prognostic value of CTCs and CTCs PD-L1.
An increasing number of studies have confirmed that genomic DNA CNV are very useful for the investigation of gene structural changes, gene expression, and tumor pathogenesis. CNV plays important roles in the development and progression of tumors, including lung cancer. In our study, sequencing results showed that all 5 samples had CNV. CNV was present in the only patient with benign disease who was CTC positive, which might have been associated with hidden cancer lesions because many studies have shown that CTCs are present in the early stage of cancer.25,26 A study in 2019 showed that the overlap between the captured CTCs and lung cancer tissue can be determined by CNV mapping of the primary tissue, metastatic lesion, and CTCs of the lung cancer patients, which further confirmed that the captured cells were tumor cells at the genomic level. The study further showed that 91% (181/198) of the mutations in CTCs of early lung cancer overlapped with the metastatic lesion after 10 months, and 79% (157/198) of the mutations overlapped with the primary lesion.27 We therefore further analyzed gene mutations. In these samples, we identified some genes related to cancer, including SMAD4, MUTYH, SKY, and ADAMTS5. SMAD4 is involved in tumor development and progression and is associated with the invasion, metastasis, and prognosis of various tumors, including lung cancer.28 MUTYH encodes a base excision repair enzyme involved in the base excision repair pathway, and a deficiency in this pathway caused by the inactivation of MUTYH protein leads to an increased gene mutation frequency and, in turn, tumor development.29 SYK is a likely potential suppressor gene and has been studied in breast cancer, pancreatic cancer, and lung cancer. In lung cancer, a deficiency of SYK expression plays important roles in angiogenesis.30,31 It has been reported that ADAMTS5 is involved in tumor metastasis and invasion and that high ADAMTS5 expression is associated with poor prognosis of lung cancer patients.32 The CTCs CNV analysis in that study showed that CTCs could be used for downstream molecule detection and that the abnormal expression of the corresponding genes was likely associated with cancer.
This study has some limitations. First, this was a single-center study with a small sample size. The patient number was small, with only 44 cases. Second, no patients showed disease progression within the short follow-up duration. Follow-up examinations of patients will be required to determine whether patients with PD-L1-positive CTCs have worse survival and whether CTCs PD-L1 expression is associated with poor prognosis. Third, the CNV analysis in the study did not have a control from lung cancer tissue and only used a negative control (Peripheral blood mononuclear cell) and a positive control from a tumor cell line (MCF7). Fourth, the number of CTCs in peripheral blood is extremely low, accounting for only 10−7 to 10−6 compared to other mono-nucleated blood cells.6 As a biomarker, a major limitation of CTCs is the low detection rate in patients with early-stage lung cancer. In the future, the CTCs detection method should be optimized. and the present technology should be standardized. A prospective study with a larger sample size and a more heterogeneous patient population is required to confirm our findings and to evaluate the benefits of this screening in the diagnosis of lung cancer.
In summary, this study confirms that the CellCollector in vivo CTCs detection technique perhaps effectively distinguish between benign and malignant nodules and maybe used for the diagnosis of early-stage lung cancer.
Acknowledgments
We acknowledge Hongxia Yang from Viroad Biotechnology Co., Ltd. for technical support and WGA and NGS data collection. This study was supported by the Hebei Provincial Key Medical Research Project (No. 20180151).
Disclosure
The authors have no conflicts of interest to declare.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39–51. doi:10.1016/j.jtho.2015.09.009
3. National Lung Screening Trial Research T, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409.
4. Patz EF
5. Wicha MS, Hayes DF. Circulating tumor cells: not all detected cells are bad and not all bad cells are detected. J Clin Oncol. 2011;29(12):1508–1511. doi:10.1200/JCO.2010.34.0026
6. Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59(1):110–118. doi:10.1373/clinchem.2012.194258
7. Patz EF
8. Cho IH, JY. Lung cancer biomarkers. Adv Clin Chem. 2015;72:107.
9. Fujiwara K, Fujimoto N, Tabata M, et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res. 2005;11(3):1219–1225.
10. Weiss G, Schlegel A, Kottwitz D, Konig T, Tetzner R. Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J Thorac Oncol. 2017;12(1):77–84. doi:10.1016/j.jtho.2016.08.123
11. Santarpia M, Liguori A, D’aveni A, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis. 2018;10(Suppl 7):S882–S897.
12. Diamandis EP, Fiala C. Can circulating tumor DNA be used for direct and early stage cancer detection? F1000Res. 2017;6:2129.
13. Zhang H, Gong S, Liu Y, et al. Enumeration and molecular characterization of circulating tumor cell using an in vivo capture system in squamous cell carcinoma of head and neck. Chin J Cancer Res. 2017;29(3):196–203. doi:10.21147/j.issn.1000-9604.2017.03.05
14. Markou A, Lazaridou M, Paraskevopoulos P, et al. Multiplex gene expression profiling of in vivo isolated circulating tumor cells in high-risk prostate cancer patients. Clin Chem. 2017;64(2):297–306. doi:10.1373/clinchem.2017.275503
15. Mandair D, Vesely C, Ensell L, et al. A comparison of CellCollector with CellSearch in patients with neuroendocrine tumours. Endocr Relat Cancer. 2016;23(10):L29–32. doi:10.1530/ERC-16-0201
16. Gorges TM, Penkalla N, Schalk T, et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin Cancer Res. 2016;22(9):2197–2206. doi:10.1158/1078-0432.CCR-15-1416
17. Li JB, Geng CZ, Yan M, et al. Circulating tumor cells in patients with breast tumors were detected by a novel device: a multicenter, clinical trial in China (in Chinese). Zhonghua Yi Xue Za Zhi. 2017;97(24):1857–1861. doi:10.3760/cma.j.issn.0376-2491.2017.24.003
18. Chen S, Tauber G, Langsenlehner T, et al. In vivo detection of circulating tumor cells in high-risk non-metastatic prostate cancer patients undergoing radiotherapy. Cancers (Basel). 2019;11(7). doi:10.3390/cancers11070933.
19. Cieslikowski WA, Budna-tukan J, Swierczewska M, et al. Circulating tumor cells as a marker of disseminated disease in patients with newly diagnosed high-risk prostate cancer. Cancers (Basel). 2020;12(1):160. doi:10.3390/cancers12010160
20. He Y, Shi J, Shi G, et al. Using the new cellcollector to capture circulating tumor cells from Blood in Different Groups of pulmonary disease: a cohort study. Sci Rep. 2017;7(1):9542. doi:10.1038/s41598-017-09284-0
21. Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res. 2009;15(22):6980–6986. doi:10.1158/1078-0432.CCR-09-1095
22. Wang X, Sun Q, Liu Q, Wang C, Yao R, Wang Y. CTC immune escape mediated by PD-L1. Med Hypotheses. 2016;93:138–139. doi:10.1016/j.mehy.2016.05.022
23. Nicolazzo C, Raimondi C, Mancini M, et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. doi:10.1038/srep31726
24. Satelli A, Batth IS, Brownlee Z, et al. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci Rep. 2016;6:28910. doi:10.1038/srep28910
25. Alix-panabieres C, Riethdorf S, Pantel K. Circulating tumor cells and bone marrow micrometastasis. Clin Cancer Res. 2008;14(16):5013–5021. doi:10.1158/1078-0432.CCR-07-5125
26. Hosseini H, Obradovic MM, Hoffmann M, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi:10.1038/nature20785
27. Chemi F, Rothwell DG, McGranahan N, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat Med. 2019;25(10):1534–1539. doi:10.1038/s41591-019-0593-1
28. Zeng Y, Zhu J, Dan S, et al. MicroRNA-205 targets SMAD4 in non-small cell lung cancer and promotes lung cancer cell growthin vitroandin vivo. Oncotarget. 2017;8(19):30817–30829. doi:10.18632/oncotarget.10339
29. Singh A, Singh N, Behera D, Sharma S. Genetic investigation of polymorphic OGG1 and MUTYH genes towards increased susceptibility in lung adenocarcinoma and its impact on overall survival of lung cancer patients treated with platinum based chemotherapy. Pathol Oncol Res. 2017;1–14.
30. Chuanliang P, Yunpeng Z, Yingtao H, Qifeng S, Xiaogang Z, Bo C. Syk expression in non-small-cell lung cancer and its relation with angiogenesis. J Can Res Ther. 2016;12(2):663–666. doi:10.4103/0973-1482.154082
31. Udyavar AR, Hoeksema MD, Clark JE, et al. Co-expression network analysis identifies Spleen Tyrosine Kinase (SYK) as a candidate oncogenic driver in a subset of small-cell lung cancer. BMC Syst Biol. 2013;7(S5):S1. doi:10.1186/1752-0509-7-S5-S1
32. Gu J, Chen J, Feng J, et al. Overexpression of ADAMTS5 can regulate the migration and invasion of non-small cell lung cancer. Tumor Biol. 2016;37(7):1–9. doi:10.1007/s13277-015-4573-x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.