Back to Journals » Journal of Blood Medicine » Volume 13
Circulating miR-126-3p and miR-423-5p Expression in de novo Adult Acute Myeloid Leukemia: Correlations with Response to Induction Therapy and the 2-Year Overall Survival
Authors Almohsen F, Al-Rubaie HA , Habib MA, Nasr SA, Perni R, Al-Quraishi L
Received 2 November 2021
Accepted for publication 20 January 2022
Published 18 February 2022 Volume 2022:13 Pages 83—92
DOI https://doi.org/10.2147/JBM.S347397
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Faez Almohsen,1 Haithem A Al-Rubaie,1 Manal A Habib,1 Sherif A Nasr,2 Rajendra Perni,2 Lubab Al-Quraishi2
1College of Medicine, University of Baghdad, Baghdad, Iraq; 2siParadigm Diagnostic Informatics, New Jersey, NJ, USA
Correspondence: Faez Almohsen, College of Medicine, University of Baghdad, Baghdad, Iraq, Tel +964 7902834062, Email [email protected]
Background: Acute myeloid leukemia (AML) results from sequential genetic alterations in a normal hematopoietic stem cell or its progenitors giving rise to an autonomous clone that dominates the bone marrow leading to marrow failure. MicroRNAs are short non-coding nucleic acid sequences that regulate post-transcriptional gene expression by base-pairing with their target mRNAs. MiRNAs can be secreted into extracellular fluids and carried to target cells by vesicles or bound to proteins. Intracellular and circulating miRNAs are believed to be useful markers in the diagnosis, prognosis, and treatment of various cancers. Practically, circulating miRNAs are more stable at room temperatures and extreme conditions.
Purpose: This study aimed to compare the expression of miR-126-3p and miR-423-5p in patients and normal subjects and correlate their expression with response to induction therapy and with their 2-year overall survival rate.
Patients and Methods: Circulating miR-126-3p and miR-423-5p was measured in the plasma of 43 adult AML patients and 35 age- and sex-matched controls by quantitative reverse transcriptase PCR. The fold change in differential expression for each gene was calculated using the comparative cycle threshold method.
Results: There was an increase in the expression of the studied miRNAs in patients compared to the control group. The average expression fold change of miR-126-3p was 3.02 (p= 0.010). The average expression fold change of miR-423-5p was 4.09 (p= 0.003). No significant correlation was found between the expression of miR-126-3p and miR-423-5p in the studied AML patients (r = 0.094, p = 0.22). Furthermore, no relationship was found between the expression of the studied miRNAs and response to induction therapy or the 2-year survival rate.
Conclusion: Although further studies are needed, our findings highlight the studied circulating miRNAs as possible diagnostic markers for AML.
Keywords: acute myeloid leukemia, miR-126-3p, miR-423-5p, response to induction therapy, overall survival, polymerase chain reaction
Introduction
Acute myeloid leukemia (AML) is known to be associated with a poor prognosis. In the USA, the 5-year survival rate of AML is 29.5%, it is estimated that 20,240 patients will be diagnosed with AML in the year 2021 and 11,400 patients will die from this disease. AML is caused by the stepwise acquisition of genetic mutations that ultimately lead to the autonomous uncontrolled proliferation and restriction of differentiation of a clone of cells which bypasses the normal physiological checkpoints, eventually leading to bone marrow failure with a consequent loss of the function of the cellular elements of blood.1,2
MicroRNAs (miRNAs) are a group of short (21–23 nucleotides) non-coding RNAs that finely regulate gene expression. A single miRNA may regulate the expression of 100 genes. Various human diseases including cancers are associated with disturbances in the miRNA expression.3,4 Most miRNAs are transcribed from their parent DNA sequences first into primary miRNAs which will then be processed into precursor miRNAs, and finally mature miRNAs. MiRNAs regulate gene expression by inducing mRNA degradation and translational repression, thereby the overexpression or the repression of miRNAs will positively or negatively regulate the degree of expression of their target genes.5
Circulating miRNAs were detected in body fluids, such as plasma and serum, urine, CSF, breast milk, colostrum, seminal fluid, tears, bronchial lavage, and peritoneal fluid. Unlike intracellular miRNA which is degraded in the extracellular environment within a few seconds, extracellular miRNAs are highly stable at room temperature for up to four days and in extreme conditions such as boiling, multiple freeze-thaw cycles, and high or low pH. This relative stability of extracellular miRNAs may be attributed to their containment within vesicles or other proteins.5–7 The unique characteristics of miRNAs like stability, tissue specificity, and testing simplicity make miRNAs encouraging targets for individualized cancer therapy. Circulating miRNAs are especially promising as rapidly emerging tools for early diagnosis, prognosis, and treatment of various cancers.
MicroRNA-126-3p (miR-126, mir-126, miRNA-126) is expressed in many human cells, such as cardiomyocytes, endothelial, and lung cells.8 The important role of miR-126-3p comes from its effect on various signaling pathways, such as CPK9 TOM1, PI3K/AKT/mTOR, and MAPK.10 MiR-126 overexpression has been associated with an increased leukemic stem cell (LSC) population and decreased normal hematopoietic stem cell counterpart.10 It was found that miR-126-3p overexpression in AML was associated with poor survival, a higher chance of relapse, and expression of genes present in AML LSC/hematopoietic stem cells (HSC) compartments.11
MicroRNA 423 was found to be upregulated in most solid tumors including gastric cancer, hepatocellular carcinoma, glioblastoma, laryngeal carcinoma, lung adenocarcinoma, breast cancer, endometrial cancer, and prostate cancer. On the other hand, miR-423-5p expression was found to be downregulated in several other tumors like ovarian cancer, osteosarcoma, and cervical cancer. The cause of this heterogeneous expression of miR-423-5p in various tumors is not fully understood and might be due to the different functional mechanisms of miR-423-5p in different tumors. The carcinogenic effect of miR-423-5p is attributed to its positive effect on cell proliferation, invasion, metastasis, and chemoresistance and its negative effect on apoptosis.12
The change in the expression of circulating miR-126-3p and miR-423-5p in AM L patients and their impact on response to induction therapy were not fully reported. Here, we aimed to find the relationship between the expression of plasma circulating miR-126-3p and miR-423-5p in adult de novo AML patients and healthy subjects and to correlate their expression with response to induction therapy, also to find the relationship between their expression and the 2-year overall survival (OS) rate.
Patients and Methods
This prospective cohort study included 43 patients with newly diagnosed de novo AML patients who attended Baghdad Teaching Hospital, Medical City from December 2018 to September 2019. This research was approved by the Research Ethics Committee at the College of Medicine, University of Baghdad, Iraq. Signed informed consent was obtained from each patient in accordance with the Declaration of Helsinki.
Based on preliminary clinical and laboratory findings supportive of an AML diagnosis, further investigations were done to patients’ peripheral blood and bone marrow specimens including (as appropriate) bone marrow aspirate and biopsy morphology, immunocytochemistry, immunohistochemistry, and flow cytometry to confirm the diagnosis and decide on the treatment strategy according to clinical and laboratory findings.
Patients were treated by a 7+3 (induction of remission) regimen. Patients were assessed for response to induction therapy on day 28. Patients were classified according to their response to induction therapy to complete remission (CR) and non-remission (NR). Patients were followed up for two years to find the 2-year OS.
Patients less than 14 years old, FAB M3 patients, patients treated by a regimen other than 7+3 regimen, treatment-related and MDS-related AML patients were excluded from this study. Thirty-five apparently healthy age- and sex-matched individuals were enrolled in this study as a control group for comparison with miRNA expression.
Platelet-poor plasma (PPP) was prepared and kept refrigerated at −80 oC. PPP samples were transferred with dry ice to si-Paradigm Diagnostic Informatics Laboratory located in New Jersey, USA, for RNA extraction, reverse transcription, and real-time PCR.
Total RNA (including miRNA) was extracted from the plasma of normal and AML patients using miRNeasy Serum/Plasma Advanced Kit (Qiagen cat#217204) and reverse transcribed with miRCURY LNA RT Kit (Qiagen cat# 339340) according to the manufacturer’s instructions. The quality of miRNA isolation, cDNA synthesis, and real-time PCR was assessed by using miRCURY LNA RNA Spike-in kit (synthetic short RNA sequences that resemble miRNAs in structure but lack close sequence similarities to known miRNAs) (Qiagen cat# 339390). The levels of expression of miR-16, miR-126-3p, and miR-423-5p were measured by quantitative RT-PCR with a miRCURY LNA SYBR Green PCR Kit (Qiagen cat# 339345) in a Qiagen Rotor-Gene Q. MiR-16-5p was used as a normalization control. The primers used for the detection of miR-126-3p, miR-423-5p, miR-16-5p, Spike in control Uni-SP2, spike-in control Uni-SP4, spike-in control Uni-SP5 and cel-miR-39-3p were the hsa-miR-126-3p miRCURY LNA miRNA PCR Assay (Qiagen cat#339306 product# YP00204227), hsa-miR-423-5p miRCURY LNA miRNA PCR Assay (Qiagen cat#339306 product# YP00205624), hsa-miR-16-5p miRCURY LNA miRNA PCR Assay (Qiagen cat#339306 product# YP00205702), Synthetic UniSP2 miRNA PCR Assay (Qiagen cat#339306 product# YP00203950), Synthetic UniSP4 miRNA PCR Assay (Qiagen cat#339306 product# YP00203953), Synthetic UniSP5 miRNA PCR Assay (Qiagen cat#339306 product# YP00203955) and miRNA cel-miR-39-3p PCR Assay (Qiagen cat#339306 product# YP00203952), respectively. All reactions were run in duplicates.
MiR-16-5p was chosen as a normalization control because it is stably expressed across normal subjects in many diseases including AML. Also, it is equal to the size of our target miRNAs, and has similar extraction efficiency and stability, as well as having expression levels within a similar range of the target miRNAs.13–15
Statistical Analyses
Regarding PCR experiments, data were acquired and analyzed by Rotor-Gene Q 2.3.5 software. The collected data were admitted into Microsoft Excel sheet 2016 and loaded into Statistical Package for the Social Sciences (SPSS) version 28 statistical program. Descriptive statistics were presented as mean ± standard deviation (SD), frequency, and percentages using tables and graphs. The Mann–Whitney U-test was used to test the statistical significance of the nonparametric variables. P values equal to or less than 0.05 were considered significant. The relative expression level was calculated by using the delta delta cycle threshold (Livak method, DDCT, or 2−ΔΔCt) analysis method.16 Pearson correlation was used for correlating miR-126-3p and miR-423-5p expression.
Results
Clinical and Laboratory Characteristics
Some clinical and laboratory characteristics of the studied patients are summarized in Table 1.
 |
Table 1 Distribution of Gender, WBC Count, Hemoglobin Concentration, Platelet Count and FAB Subtype in the Studied Patients |
 |
Table 2 Mean, Median and Standard Deviations (SD) of the Studied miRNA PCR Values (Ct, Δct, Δδct and Fold Change) |
MicroRNA Expression
The expressions of miR-126-3p and miR-423-5p were assayed in 43 AML patients and 35 normal controls using qRT-PCR. The mean relative Ct (∆Ct), ∆∆Ct and fold change of miR-126-3p and miR-423-5p in patients and controls are shown in Figure 1 and Table 2.
 |
Figure 1 Comparison between the average fold change (2−ΔΔCt) of miR-126-3p and miR-423-5p in patients and control groups (p=0.010 and 0.003, respectively). |
Correlation Between miR-126-3p and miR-423-5p Expression
Using Pearson correlation for linear regression analysis, no significant correlation was found between the expression of miR-126-3p and miR-423-5p in the studied AML patients, with a correlation coefficient r of 0.094 and p-value of 0.22 (Figure 2).
 |
Figure 2 Correlation between miR-126-3p and miR-423-3p expression in the studied patients represented as pairs. |
Relationship Between miR-126-3p and miR-423-5p Expression and Response to Induction Therapy
The mean relative expression (fold change) of miR-126-3p in the CR group was 2.11 ± 2.03 (range 0.29–7.90, median = 1.54), whereas the mean relative expression in the NR group was 3.91 ± 4.16 (range 0.11–19.73, median = 3.49), p value (Mann Whitney test) was 0.151, showing no significant statistical difference.
The mean fold change of miR-423-5p in the CR group was 3.26 ± 2.27 (range 0.00–9.47, median = 3.56), whereas the mean fold change in the NR group was 4.04 ± 4.36 (range 0.00–13.82, median = 2.03), p value (Mann Whitney test) was 0.621 showing no significant statistical difference (Figure 3).
 |
Figure 3 MiR-126-3p and miR-423-3p expression in the CR and NR groups. |
Relationship Between miR-126-3p and miR-423-5p Expression and the 2-Year OS
Out of 37 patients who could be tracked up to 2 years, only five could survive 2 years or more after diagnosis. The mean relative expression (fold change) of miR-126-3p in the 2-year survivor group was 2.36 ± 2.56 (range 0.28–6.55, median = 1.43), whereas the mean relative expression in the 2-year non-survivor group was 3.22 ± 3.68 (range 0.08–19.73, median = 2.32), p value (Mann Whitney test) was 0.560, showing no significant statistical difference. The mean relative expression (fold change) of miR-423-5p in the 2-year survivor group was 2.72 ± 3.10 (range 0.00–7.71, median = 1.72), whereas the mean relative expression in the 2-year non-survivor group was 4.28 ± 5.22 (range 0.00–22.7, median = 3.30), p value (Mann Whitney test) was 0.589, showing no significant statistical difference (Figure 4).
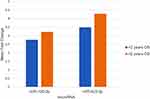 |
Figure 4 Comparison between miR-126-3p and miR-423-5p mean fold change of 2-years survivors (blue) and non-survivors (red). |
Discussion
Despite decades of research in the various aspects of AML, including new therapeutic approaches and prognostic markers, most patients do not survive more than 5 years. To improve this outcome, more in-depth study and utilization of the currently available and newly discovered markers should be done. MicroRNAs are relatively novel players that have been dysregulated in many diseases including cancers. In addition, they have been proposed as possible therapeutic targets and markers for the prediction of outcome. Circulating miRNAs can be secreted by many cancer cells including leukemic cells and can be promising markers in the diagnosis and prognosis of AML. To our knowledge, the study of circulating miRNA-126-3p and miR-423-5P in AML patients’ plasma was not done by any researcher before.
In this study, a significant increase in miR-126-3p expression in the studied AML patients was observed. Although the expression level of plasma miR-126-3p was higher in the NR group than in the CR group, this was not statistically significant.
MiR-126-3p is a short non-coding RNA with a wide range of effects, through its regulatory role of several signaling molecules. MiR-126-3p down-regulates CT10 regulator of kinase (CRK), an intracellular signal transduction molecule important in regulating cell adhesion, proliferation, migration, and invasion.9 Dysregulation of the CRK pathway is associated with increased susceptibility to cancer.17 MiR-126-3p upregulates CXCL12 expression.18 Interaction between CXCL12 expressed on stromal cells and its receptor CXCR4 (CD184) on leukemic blasts is important to keep leukemia homed to its microenvironment protected from chemotherapy, also it counteracts apoptosis and recruits progenitor cells.19 High CXCR4/CXCL12 expression was associated with adverse prognosis and has been a potential target for anti-leukemia drugs.20,21 MiR-126-3p downregulates PU.1, a transcription factor involved in the differentiation of myeloid cells and compromised in several AML subtypes. Minimal reduction of PU.1 was linked to leukemia initiation and the formation of preleukemic stem cells in AML.22 It has been found that miRNA-126-3p is downregulated in several solid tumors.23 In AML, Li et al found that miRNA-126-3p was upregulated in AML with inv(16)/CBFβ–MYH11 or t(8;21)/AML1-ETO.24
MiR-126-3p is also proposed to exhibit its leukemogenic properties by targeting the PI3K/AKT/mTOR signaling pathway to promote quiescence, increase self-renewal, and promote chemotherapy resistance of LSC.25 PI3K/AKT/mTOR signaling is overactive in many cancers including AML, thus reducing apoptosis, and allowing proliferation, giving a proliferation advantage over differentiation.26–28 Therapeutic agents to target PI3K/AKT/mTOR signaling pathway have been studied. Dual PI3K/mTOR inhibition was cytotoxic not only to leukemic blasts’ bulk but also to leukemia-initiating cells.29
MiRNA-126 has been studied as a potential therapeutic target in AML. Targeting of miRNA-126 LSCs by nanoparticles containing antagomiR-126 resulted in a reduction of LSCs by depletion of the quiescent cell subpopulation.30 In addition, therapeutic depletion of miR-126 in inv(16) AML xenograft models with a specifically designed inhibitor of miR-126 called miRisten inhibited the survival of AML cells, reduced leukemia burden and LSC activity.31
In this study, circulating miR-423-5p was significantly overexpressed in AML; however, there was no statistically significant relationship between plasma miR-423-5p expression and response to induction therapy in the studied AML patients.
The study of miR-423 in a normal state and different solid tumors revealed its effects on various processes inside the cells and its role in the biology of solid tumors. However, its role in leukemias has not been fully elucidated. MiR-423 negatively regulates colony-stimulating factor 1 (CSF1), a cytokine that controls the production, differentiation, and function of macrophages, and may affect CDK expression and induce cell arrest in the G0/G1 phase, by upregulating Cyclin-dependent kinase inhibitor 1A (CDKN1A), a negative regulator of the cell cycle, keeping the cell in a quiescent state, a requirement for LSCs to escape the effect of chemotherapeutic drugs and to keep a reserve of leukemia-initiating cells.32
Overexpression of miR-423-3p indicated poor prognosis and promoted cell proliferation, migration, and invasion in lung cancer.33 Metastasis suppressor 1 (MTSS1), a multifunctional molecular effector with an important role in cancer development, carcinogenesis, and metastasis, was a direct miR-423-5p target.34,35 It was observed that miR-423-5p mimics promoted the proliferation and inhibited apoptosis of bone marrow mesenchymal stem cells from orofacial bone (OMMSCs), while antisense inhibitors of this miRNA had the opposite effect.36 In the setting of leukemia, very few studies relevant to miR-423 were found. A study of bone marrow circulating miRNAs in AML found that miR-423-5p was overexpressed in AML bone marrow samples.37 Moreover, MiR-423 (among other miRNAs) was overexpressed in two AML cell lines HL-60 (human promyelocytic leukemia cell line) and THP-1 (human monocytic leukemia cell line).38 Merkerova et al studied the miRNA expression profile of 1145 miRNAs in CD34+ cells of MDS patients and found that hsa-miR-423-5p was downregulated in those patients.39
Conclusion
There was an increase in the expression of circulating miR-126-3p and miR-423-5p in AML patients as compared to normal age- and sex-matched controls. This might highlight the role of both miRNAs in the initiation of AML and the possibility of being diagnostic and therapeutic targets. This is the first report of the significance of the plasma circulating form of both miRNAs in AML patients, which might pave the way for further studies with a larger cohort of patients to compare the plasma levels of the studied miRNAs with other diagnostic and prognostic indicators in AML, given that plasma miRNAs are stable at extreme conditions and can be measured with relative ease compared to intracellular miRNAs.
Data Sharing Statement
Data related to this research paper including raw data, materials and methods, PCR curves, Spike-in control data, software and algorithms will be provided upon reasonable request.
Ethics and Consent
This research was approved by the Research Ethics Committee at the College of Medicine, University of Baghdad, Iraq. Signed informed consent was obtained from each patient in accordance with the Declaration of Helsinki. There is no material reproduced from other sources to need special permission from the copyright owner. This research does not include any clinical trial.
Acknowledgments
We are thankful to the staff of the hematology laboratory and the clinical hematology department of the Medical City complex for their cooperation in the stage of sample collection. Also, we are grateful to the staff of siParadigm Diagnostic Informatics for providing full assistance in performing PCR experiments of this work.
Funding
This work was fully funded by Sherif A Nasr, president of siParadigm Diagnostic Informatics clinical laboratory located in New Jersey, USA, where all PCR experiments were done.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech. 2014;7(8):941–951. doi:10.1242/dmm.015974
3. Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol. 2010;10(5):543–550. doi:10.1016/j.coph.2010.05.010
4. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D62. doi:10.1093/nar/gky1141
5. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. doi:10.3389/fendo.2018.00402
6. Sohel MH. Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achievements Life Sci. 2016;10(2):175–186. doi:10.1016/j.als.2016.11.007
7. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi:10.1073/pnas.0804549105
8. Nikolic I, Plate KH, Schmidt MHH. EGFL7 meets miRNA-126: an angiogenesis alliance. J Angiogenes Res. 2010;2(1):9. doi:10.1186/2040-2384-2-9
9. Feng R, Chen X, Yu Y, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi:10.1016/j.canlet.2010.06.004
10. Zhang Y, Huang B, Wang HY, Chang A, Zheng XFS. Emerging role of MicroRNAs in mTOR signaling. Cell Mol Life Sci. 2017;74(14):2613–2625. doi:10.1007/s00018-017-2485-1
11. de Leeuw DC, Denkers F, Olthof MC, et al. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014;74(7):2094–2105. doi:10.1158/0008-5472.CAN-13-1733
12. Ke R, Lv L, Zhang S, Zhang F, Jiang Y. Functional mechanism and clinical implications of MicroRNA-423 in human cancers. Cancer Med. 2020;9(23):9036–9051. doi:10.1002/cam4.3557
13. Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381. doi:10.1136/gut.2008.167817
14. miRCURY LNA miRNA PCR Handbook. Qiagen [Internet]; October, 2019. Available from: https://www.qiagen.com/us/resources/resourcedetail?id=f02862d7-29df-4466-a457-ebde78beb891&lang=en.
15. Fayyad-Kazan H, Bitar N, Najar M, et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med. 2013;11:31. doi:10.1186/1479-5876-11-31
16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
17. Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi:10.1186/1478-811X-7-13
18. Bassand K, Metzinger L, Naim M, et al. miR-126-3p is essential for CXCL12-induced angiogenesis. J Cell Mol Med. 2021;25(13):6032–6045. doi:10.1111/jcmm.16460
19. Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi:10.1126/scisignal.2000610
20. Cho BS, Kim HJ, Konopleva M. Targeting the CXCL12/CXCR4 axis in acute myeloid leukemia: from bench to bedside. Korean J Intern Med. 2017;32(2):248–257. doi:10.3904/kjim.2016.244
21. Yazdani Z, Mousavi Z, Moradabadi A, Hassanshahi G. Significance of CXCL12/CXCR4 ligand/receptor axis in various aspects of acute myeloid leukemia. Cancer Manag Res. 2020;12:2155–2165. doi:10.2147/CMAR.S234883
22. Will B, Vogler TO, Narayanagari S, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015;21(10):1172–1181. doi:10.1038/nm.3936
23. Li XM, Wang AM, Zhang J, Yi H. Down-regulation of miR-126 expression in colorectal cancer and its clinical significance. Med Oncol. 2011;28(4):1054–1057. doi:10.1007/s12032-010-9637-6
24. Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105(40):15535–15540. doi:10.1073/pnas.0808266105
25. Lechman ER, Gentner B, Ng SWK, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29(4):602–606. doi:10.1016/j.ccell.2016.03.015
26. Qiu HY, Wang PF, Zhang M. A patent review of mTOR inhibitors for cancer therapy (2011-2020). Expert Opin Ther Pat. 2021;1–11. doi:10.1080/13543776.2021.1940137
27. Pappa T, Ahmadi S, Marqusee E, et al. Oncogenic mutations in PI3K/AKT/mTOR pathway effectors associate with worse prognosis in BRAF(V600E)-driven papillary thyroid cancer patients. Clin Cancer Res. 2021;27(15):4256–4264. doi:10.1158/1078-0432.CCR-21-0874
28. Li H, Zhao S, Shen L, et al. E2F2 inhibition induces autophagy via the PI3K/Akt/mTOR pathway in gastric cancer. Aging (Albany NY). 2021;13(10):13626–13643. doi:10.18632/aging.202891
29. Bertacchini J, Frasson C, Chiarini F, et al. Dual inhibition of PI3K/mTOR signaling in chemoresistant AML primary cells. Adv Biol Regul. 2018;68:2–9. doi:10.1016/j.jbior.2018.03.001
30. Dorrance AM, Neviani P, Ferenchak GJ, et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29(11):2143–2153. doi:10.1038/leu.2015.139
31. Zhang L, Nguyen LXT, Chen YC, et al. Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance. Nat Commun. 2021;12(1):6154. doi:10.1038/s41467-021-26420-7
32. Xie S, Zhang Q, Zhao J, Hao J, Fu J, Li Y. MiR-423-5p may regulate ovarian response to ovulation induction via CSF1. Reprod Biol Endocrinol. 2020;18(1):26. doi:10.1186/s12958-020-00585-0
33. Wang RLG, Zhuang G, Sun S, Song Z, Song Z. Overexpression of microRNA-423-3p indicates poor prognosis and promotes cell proliferation, migration, and invasion of lung cancer. Diagn Pathol. 2019;14(1). doi:10.1186/s13000-019-0831-3
34. Sun G, Ding X, Bi N, et al. MiR-423-5p in brain metastasis: potential role in diagnostics and molecular biology. Cell Death Dis. 2018;9(10):936. doi:10.1038/s41419-018-0955-5
35. Ling DJ, Chen ZS, Liao QD, Feng JX, Zhang XY, Yin TY. Differential effects of MTSS1 on invasion and proliferation in subtypes of non-small cell lung cancer cells. Exp Ther Med. 2016;12(2):1225–1231. doi:10.3892/etm.2016.3382
36. Yang S, Guo J, Zhou L, Xing H, Wang X, Dong C. miR-148b-3p, miR-337-5p and miR-423-5p expression in alveolar ridge atrophy and their roles in the proliferation and apoptosis of OMMSCs. Exp Ther Med. 2018;16(6):5334–5342. doi:10.3892/etm.2018.6850
37. Moussa Agha D, Rouas R, Najar M, et al. Identification of acute myeloid leukemia bone marrow circulating MicroRNAs. Int J Mol Sci. 2020;21(19):7065. doi:10.3390/ijms21197065
38. Xiong Q, Yang Y, Wang H, et al. Characterization of miRNomes in acute and chronic myeloid leukemia cell lines. Genomics Proteomics Bioinformatics. 2014;12(2):79–91. doi:10.1016/j.gpb.2014.02.001
39. Dostalova Merkerova M, Krejcik Z, Votavova H, Belickova M, Vasikova A, Cermak J. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. Eur J Hum Genet. 2011;19(3):313–319. doi:10.1038/ejhg.2010.209
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
