Back to Journals » Cancer Management and Research » Volume 13
Circular RNAs: A Promising Biomarker for Endometrial Cancer
Authors Guo J , Tong J , Zheng J
Received 7 November 2020
Accepted for publication 19 January 2021
Published 17 February 2021 Volume 2021:13 Pages 1651—1665
DOI https://doi.org/10.2147/CMAR.S290975
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Eileen O'Reilly
Jialu Guo, 1, 2 Jinyi Tong, 1, 2 Jianfeng Zheng 2, 3
1Department of the Fourth Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310008, People’s Republic of China; 2Department of Obstetrics and Gynecology, Hangzhou Women’s Hospital (Hangzhou Maternity and Child Health Care Hospital), Hangzhou, Zhejiang Province, 310008, People’s Republic of China; 3Department of Obstetrics and Gynecology, Affiliated Hangzhou Hospital, Nanjing Medical University, Hangzhou, Zhejiang Province, 310008, People’s Republic of China
Correspondence: Jinyi Tong
Department of the Fourth Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou Women’s Hospital (Hangzhou Maternity and Child Health Care Hospital), Hangzhou, Zhejiang Province, 310008, People’s Republic of China
Tel +86 13588818180
Email [email protected]
Abstract: Endometrial cancer (EC) is one of the most common malignant tumors of the female reproductive tract. EC patients have high morbidity and mortality rates and remain an important cause of cancer-related morbidity and mortality worldwide. More and more studies have shown that a large number of non-coding RNAs (such as microRNAs and long non-coding RNAs) are associated with the occurrence of diseases. Circular RNAs (circRNAs) is an endogenous non-coding RNA. It has a unique covalent structure. Many studies in recent years have found circRNAs differential expression in a variety of tumor tissues compared to matched normal tissues. In endometrial carcinoma, there also are multiple circRNAs differentially expressed and therefore circRNAs perhaps can be used as a diagnostic and prognosis biomarkers of EC. In this review, we described the biogenesis, function and characteristics of circRNAs, and the circRNAs with potential influence and clinical significance on the development of EC were summarized. Adenocarcinoma is the most common form of EC, so this review focuses on endometrioid adenocarcinoma.
Keywords: circRNA, biogenesis, back-splicing, function, endometrial cancer, biomarker
Background
Endometrial cancer (EC) is one of the most common malignant tumors of the female reproductive tract. Each year, approximately 142,000 women worldwide develop endometrial cancer and an estimated 42,000 women die of this cancer.1 It is the fourth most common cancer among women in the United States, after breast, lung, and colorectal cancers.2 Most cases of EC are diagnosed after menopause and the highest incidence rate is around 70 years old. Estrogen therapy, early menstruating, late menopause, tamoxifen therapy, infertility, polycystic ovary syndrome, increased age, obesity, hypertension, diabetes, and hereditary nonpolyposis colorectal cancer can all be risk factors.3 Survival is usually determined by the stage and histology of the disease, and the prognosis of endometrial cancer varies greatly in different stages and histological types. The most common lesions (type I) are typically hormone-sensitive and low-stage and have a good prognosis, while type II tumors have a high grade and are prone to relapse even in the early stages.4 The majority of type I EC are endometrioid carcinoma. The main treatments for EC are total hysterectomy and bilateral salpingo-oophorectomy. Radiation and chemotherapy can also play a role in treatment.3 However, morbidity and mortality rates among patients with EC remain high and EC remains an important cause of cancer-related morbidity and mortality globally. Studies have shown that a large number of non-coding RNAs (such as microRNAs and long non-coding RNAs) are associated with the occurrence of gynecological diseases.5–9 Recently, some studies aimed to investigate the expression and function of circRNAs in EC.10,11
CircRNAs are the latest members of a growing world of RNA molecules. Different from classical RNA formation, circRNAs are formed by back-splicing, a special splicing method, in which the 5 ‘-end of more than one exon or intron is covalently connected to the 3ʹ -end to form circular RNA molecules.12,13 With the wide application of gene sequencing technology and bioinformatics methods, many studies have shown that circRNAs are abnormally expressed in many malignant tumors.14–16 Due to the covalent ring structure of circRNAs, they are not easy to be degraded by exonuclease, and because of their tissue stability and specificity, more and more circRNAs have been confirmed as diagnostic and prognostic biomarkers for various diseases.
Therefore, circRNAs may be used as diagnostic and therapeutic biomarkers for EC. In this review, we describe recent research progress of circRNAs in the biogenesis and function of endometrioid adenocarcinoma, so as to prepare for further research on the application of circRNAs in EC diagnosis, prognosis and treatment.
Biogenesis of circRNAs
In all eukaryotes, the removal of introns and linking exons is a major part of the RNA splicing process, transforming the precursor mRNAs (pre-mRNAs) containing introns into intron-less mRNAs. Most eukaryotic circRNAs are produced by pre-mRNAs. In recent years, another common splicing method has been found in eukaryotes. It can covalently connect the 5 ‘-end of more than one exon or intron to the 3ʹ -end to form a circular RNA molecule. This splicing method is called back-splicing.12,17
The splicing regulatory mechanisms of circRNAs biogenesis are different from the linear isoforms. CircRNAs demonstrate significant and diverse back-splicing events catalyzed by typical spliceosome mechanisms in different cell lines.13,18–20 CircRNAs are pervasive in molecular biology. According to splicing mode, circRNAs can be divided into four types: exonic circRNAs (ecRNAs), exon-intron circRNAs (EIciRNAs), intronic circRNAs (ciRNAs) and intergenic circRNAs (IciRNAs).21–23 So far, six biogenesis mechanism of circRNAs have been proposed, including lariat-driven circularization, intron pairing-driven circularization, RNA-binding proteins (RBPs)-mediated circularization, direct circularization of lariat introns, tRNA splicing-driven circularization, and ribosomal RNA (rRNA) splicing-driven circularization.24–27This review focuses on the first three mechanisms.
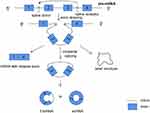
The first biogenesis mechanism of circRNAs is lariat-driven circularization. This model that leads to back-splicing is exon skipping, in which one or more exons are missing from the mature mRNAs. The 3ʹ -end splicing ligands of exons covalently bind to the 5ʹ -end splicing receptors of exons, and then the introns are excised to form circRNAs. In this model, the lariat-driven circularization is formed by the connection of two non-adjacent exons, and finally producing a lariat structure, a mRNA with skipped exons and a circular RNA transcript. The exon-skipping events produce an exon-containing lariat, which could then itself be internally spliced to an exon circle (Figure 1). In other words, exon-skipping leads to a lariat, whose restricted structure promotes circularization.28–30 Moreover, intronic lariats can form intronic circRNAs (ciRNAs) (Figure 2). Intron cyclization mainly exists in the nucleus, and their formation depends on 7 nucleotide GU enrichment elements containing adjacent 5ʹ splicing sites and 11 nucleotide C enrichment elements containing adjacent branch sites, with a small number of miRNA targets.27,31
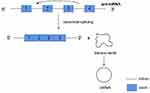 |
Figure 2 Intronic lariats can form intronic circRNAs (ciRNAs). |
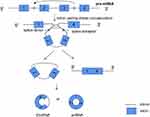
The second biogenesis mechanism of circRNAs is intron pairing-driven circularization. Exons of the former mRNA or two introns on either side of the exon that can be connected to each other. The flanking introns are close to each other, forming a secondary conformation, which enables the splicing site to undergo back-splicing (Figure 3). Most of the intron-pairing patterns are facilitated by ALU repetition.32 In other words, the two non-adjacent introns were first paired with each other to form a ring structure, then the introns were excised, and finally circRNA was formed. From the first model we can see that, ecRNA can be formed by lariat-driven circularization. Alternatively, ecRNA could also be formed by alternative 5′ to 3′ splicing of nascent transcripts.28
The third circRNAs formation mechanism is by RNA-binding proteins (RBPs). This mechanism involves the ability of protein factors that bind to the former mRNA to link flanking introns, a process induced by protein dimerization that produces RNA rings (Figure 4). And muscleblind like splicing regulator 1 (MBNL1) protein is one of the most popular RBPs responsible for circRNA biogenesis. MBNL1 was shown to bind to its own pre-mRNA and binds the two flanking introns together.13 CircRNA MBL/MBNL1 itself contains conservative muscleblind (MBL) binding sites, so it is easy to bind to the MBL protein. This binding effect promotes the biosynthesis of circMBL, and the MBL level is important to determine the cyclization rate of bracketed exons. MBL expression may reduce the production of parental mRNA by promoting circMBL production.13,33–35 Another RNA-binding proteins (RBPs), adenosine deaminase 1 acting on RNA (ADAR1), have been reported to play a role in circRNAs biogenesis. The downregulation of ADAR1 specifically up-regulated the expression of some circRNA, suggesting that ADAR1 plays a role in inhibiting the biogenesis of circRNA.32,34 This regulation is associated with Adenosine-to-Inosine (A-to-I) editing.32 Double stranded RNA (dsRNA) paired structures are called A-to-I RNA editing targets by ADARs.36,37 Under normal conditions, high-enrichment A-to-I editing of dsRNA regions can reduce RNA pairing structure, resulting in a reduction in RNA pairing and thus a reduction in the efficient back-splicing of circRNAs formation. However, with the decrease of A-to-I editing level after ADAR1 gene knockout, the pairings of RNA to cross-introns became more stable, which was conducive to back-splicing to produce circRNAs.32
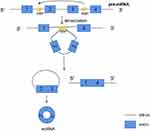 |
Figure 4 RNA binding proteins (RBP) bind to introns on both sides of the exon that forms circRNA. RBP dimerization promotes the back splicing process. |
Function of CircRNAs
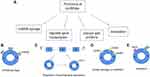
CircRNAs are rich and evolutionarily conserved RNAs of largely unknown function. CircRNAs have a wide range of biological functions. Here, we introduce some of its functions (Figure 5A). This is described further below.
CircRNA Act as miRNA Sponge (Figure 5B)
The most common function of circRNAs is the miRNA sponge. MicroRNAs (miRNAs) are another type of non-coding RNA that is transcribed from the precursors of short hairpins.38 CircRNAs can act as microRNA (miRNA) sponge by binding to miRNA and inhibiting its function, thereby releasing downstream target genes from miRNA-mediated repression and inhibiting the ability of miRNA to perform its post-transcriptional repression. MiRNAs are important post-transcriptional regulators of gene expression and play their roles through direct base pairing with the target sites in the non-translational region of messenger RNAs. And the activity of miRNAs has been shown to be influenced by the presence of miRNA sponge transcripts, so-called competitive endogenous RNAs in humans and target imitations in plants.39–41 miRNA sponges are widespread regulators of miRNA activity in many eukaryotes. CircRNA is a highly prevalent RNA species in human transcripts.42 Many abundant endogenous circRNAs molecules are naturally resistant to the decline of extracellular dissolved RNA and can be used as effective miRNA sponges to increase the growing lineage of gene expression regulation.
Antisense to the cerebellar degeneration-related protein 1 transcript (CDR1as) and the testis-specific circRNA, Sex-determining region Y(Sry) are the most representative miRNA sponges.40,43,44 The exonic circRNAs of CDR1as and Sry have been shown to bind miRNAs without being degraded, making them excellent candidates for competing endogenous RNA activity.12 CDR1as acts as a microRNA-7 (miR-7) sponge, therefore, we also term this circular transcript ciRS-7 (circular RNA sponge for miR-7). And Sry serves as a miR-138 sponge. Nowadays, many studies have found extensive involvement of miR-7 as a key regulator of many cancer pathways, so CIRS-7 may be an important factor associated with cancer.45,46
CircRNA Regulates Gene Transcription (Figure 5C)
CircRNAs constitute a transcription family with unique structures and still largely unknown functions. Because the source of introns circRNA (ciRNA) contains very little of microRNAs binding sites, and the small amount of binding sites is more dispersed, so they are not ordinary type circRNA have the role of miRNA sponges, but ciRNA has positive regulation of RNA polymerase II transcriptional activity function.27,47
Li et al revealed a new role for circRNAs in regulating gene expression in the nucleus. Some EIciRNA regulate the transcription of parental genes by interacting with U1 small nuclear RNA (snRNP). First, EIciRNA and U1 small nuclear ribonucleoprotein (snRNP) form the EIciRNA-U1 snRNP complex, then the compound with RNA polymerase II transcription complex interaction to promote gene expression.48 This mode of RNA-RNA interaction to regulate transcription not only enriches the functions of circRNAs, but also provides a new direction for the research of circRNAs.
CircRNA Interacts with Proteins (Figure 5D)
CircRNAs can interact with proteins and thus affect the function of proteins. CircRNAs can stably bland to AGO proteins and RNA polymerase II.28,49 CDR1as and Sry in exonic circRNA bind to the miRNA effector Argonaute (AGO).27,49 In addition, they can be combined with a variety of RBPs to act as RBP scaffolds.12 CircRNA may also be used as a target sequence element to simultaneously bind RBP, RNA or DNA using its complementary sequences.49 CDR1as contains 74 miR-7 seed sequence matches and is tightly bound by Argonaute proteins (the proteins that bind to miRNAs).12,49 Similarly, when miR-138 is overexpressed, the circular Sry transcript has 16 binding sites for miR-138 and co-precipitates with Argonaute 2 (AGO2).40
Ankyrin repeat domain 52 circular intronic RNA (ci-ankrd52), eukaryotic translation initiation factor 3 subunit J circular RNA (circEIF3J) and poly-adenylate-binding protein-interacting protein 2 circular RNA (circPAIP2) can interact with the RNA polymerase II complex and ultimately regulate transcription.27
Ashwal-Fluss et al found that the circMBL can bind to RBPs. When the cells contain excessive MBL protein, circMBL will be promoted to reduce the mRNA production of the protein, while circMBL will bind to the excessive MBL protein, making the MBL protein content tend to be normal.13
Du et al found that circ-Foxo3 can stay in the cytoplasm through interactions with anti-aging and stress protein-related factors inhibitor of DNA-binding 1 protein (ID-1), focal adhesion kinase (FAK), and hypoxia inducible factor 1α (HIF1α) in humans, thereby inhibiting the corresponding resistance process.50 The circRNAs can also form the circ-Foxo3-P21-CDK2 ternary complex, inhibit the function of cyclin- dependent kinase 2(CDK2) and block the cell cycle process.51
CircRNA is Involved in Protein Translation (Figure 5E)
If circRNA contains internal ribosomal entry sites (IRES), it may guide protein synthesis. Eukaryotic ribosome 40s small subunits can enter the IRES of these circRNA to guide protein synthesis.52 It turns out that if circRNA contains bases that are multiples of 3, the circRNA encoding the protein circulates around the reading frame, translating a repeated polypeptide sequence. Some studies have showed that some circRNAs carry open reading frames, and can be translated into peptides or proteins.53,54 The presence of IRES has been demonstrated that circRNA translation is driven by a cap-independent mechanism.55
So far it’s been found that circRNA of hepatitis delta virus (HDV) and circRNA in rice yellow mottle virus can code proteins.56,57 HDV contains a single stranded circular RNA molecule. This is the first animal virus to be identified as a circRNAs genome. CircRNAs have only been found in plant viruses.58 Peptides or proteins encoded by circRNAs play a key role in mediating cancer development. Therefore, it seems necessary to investigate whether proteins translated from circRNA have function in gynecological cancers.59,60
General Characteristics of CircRNAs
1. Unlike linear RNA, circRNA forms a covalent circular structure, with no 5′ end caps or 3′poly(A) tails, which is not easy to be degraded by exonuclease and is more stable than linear RNA.61
2. CircRNAs are a wide variety, and some are more abundant than their linear mRNA analogues.26,62 CircRNA was formed from introns, exons, and intergene regions, and even 50 and 30 untranslated fragments.27,49,63
3. CircRNAs are mainly composed of exons and a small part of circRNA is formed by intron cyclization.
4. Some circRNAs, rich in microRNA response elements (MRE), can play the role of competitive endogenous RNAs, bind with miRNAs, play the role of miRNA sponge in cells, and inhibit the function of miRNAs so as to regulate gene expression levels.40
5. CircRNAs are expressed in tissue-specific, cell-specific, and developmental stage-specific patterns.64,65 It is dynamically expressed during development and expressed in a tissue-specific manner.48,49,66,67
6. Most circRNAs are endogenous ncRNA, but only some exogenous circRNAs can be translated and expressed, such as hepatitis delta virus (HDV) and engineered circRNA with internal ribosome entry site (IRES).52,68
7. CirRNAs are evolutionarily conservative between different species.28,49,66,69
8. With the exception of intron-containing circRNA mainly present in the nucleus, most circRNAs are exported to the cytoplasm in a size-dependent manner after biogenesis.70
These characteristics of circRNA suggest that it may play an important role at both transcriptional and post-transcriptional levels and may be an ideal marker for disease diagnosis.
Circular RNAs and Endometrial Cancer (EC)
More and more studies have confirmed abnormal circRNAs expression in multiple tumor tissues compared with matched normal tissues. For example, many researches have studied the relationship between circRNAs and cervical cancer, and the results show that circRNAs are involved in the development of cervical cancer through various mechanisms, among which the spongification of miRNA is the most important one. Studies have shown that has_circ_0018289, has_circ_0001445, has_circ_0023404, has_circ_0000263, circRNA-000284, circRNA8924, has_circRNA_101996, circ-ATP8A2, circ_0067934 and circEIF4G2 are abnormally expressed in cervical cancer cells.71–76 In addition, in ovarian cancer, circHIPK3 was found to be highly expressed in epithelial ovarian cancer (EOC) and ovarian cancer cells A2780, HO-8910, SKOV3 and CAOV3.77
Endometrial Cancer (EC)
Endometrial cancer is a group of malignant epithelial neoplasms occurring in the endometrium. It usually occurs in perimenopausal and postmenopausal women. Endometrial cancer is one of the most common tumors of the female reproductive system and the third most common gynecologic malignancy leading to death (second only to ovarian cancer and cervical cancer). The disease is closely related to lifestyle. And the incidence varies from region to region. EC is rare in less developed countries because of fewer risk factors, but specific mortality rates are higher.78,79 The incidence rate in North America and Europe is 10 times higher than that in less developed countries. In North America and Europe, the incidence rate is second only to breast cancer, lung cancer and colorectal cancer, and it ranks the first among female reproductive system cancers.2,80
EC is generally classified into type I and type II according to histological type.4 Type I EC are usually low stage and low grade, positive for estrogen and progesterone receptors, and has a good prognosis. In contrast, type II EC is usually estrogen-independent and has a poor prognosis.10,81 The majority of type I EC are endometrioid carcinoma and a few were mucinous adenocarcinoma. Estrogen-independent endometrial carcinoma includes serous carcinoma, clear cell carcinoma and so on.1 The most common histological subtype of EC is endometrioid adenocarcinoma,81 which is also main described in this review. Endometrioid carcinoma is classified into three levels according to the degree of cell differentiation or the proportion of solid components. It is highly differentiated (G1), moderately differentiated (G2) and poorly differentiated (G3), and the malignant degree of poorly differentiated tumor is high.1,82
The incidence of EC increases with the increase in life expectancy. Age-adjusted morbidity increased even with hysterectomy.83 The rise is linked to an epidemic of obesity and physical inactivity.84 EC is one of the few human malignancies with rising mortality rates,85 highlighting the urgent need to develop more effective strategies to diagnose and treat the disease. To explore the relationship between circRNAs and EC is helpful for further understanding and treatment of EC.
Expression of CircRNAs in Endometrial Cancer
CircRNAs have covalently closed structure and are more stable than other RNAs. This stability may prove to be an ideal property for circRNA in their future development as biomarkers.14,86–88 CircRNAs have also been shown to be useful molecules in the treatment of a variety of diseases, including neurological disorders, cardiovascular disease, cancer and so on.65,89–91 The role of circRNAs in cancer pathology has recently been the subject of numerous studies.92–94 CircRNAs have been linked to many cancers. CircRNAs from tumor suppressor gene FBXW7 could be translated into protein products that reduced the half-life of c-Myc.95 There are also two types of circRNAs, circHIPK3 and circDOCK1, regulate cell growth and act as cancer biomarkers.96,97 For example, in colorectal cancer and ovarian cancer, the abundance of circRNAs, as measured by the ratio of circRNAs to linear isoforms, is lower in tumor samples.98 And the ratio was negatively correlated with the proliferation rate of tumor cells. In addition, compared with healthy controls, peripheral blood exosome circRNAs showed unique expression patterns in colon cancer patients.99 The functional significance of these circRNAs in cancer is not fully established, but they may serve as potential biomarkers for EC diagnosis or developmental monitoring. Although there is increasing evidence that circRNAs play a role in tumorigenesis and cancer progression, their role in EC is completely unknown.
So far, few studies have been published on circRNAs in EC. The expression profile of circRNAs in EC tissues was changed compared with that in adjacent normal tissues.10 Similarly, studies have confirmed that circRNAs expression in grade 3 EC tissues is significantly different from that in adjacent non-cancerous endometrium, which may provide new molecular candidates for the diagnosis and clinical treatment of grade 3 EC.11 The overall abundance of circRNAs in EC was lower than normal endometrium. There was no difference in the number of transcripts between EC and normal endometrium for linear RNA. In addition, there are many hotspot genes for circRNAs transcription that may account for changes in circRNAs expression between normal and malignant endometrium.10
One study listed the top 10 unique hotspot genes expressed in normal tissue and 8 unique hotspot genes expressed in EC tissue:DNAH14, MT-RNR2, RABGAP1, ESR1, FIP1L1, GFPT1, INADL, PCNX.10 Hotspot genes are defined as the production of more than a dozen different circRNA isoforms in a given tissue or cell.100 Some studies have found that the expression changes of circRNAs in EC are the result of the expression changes of specific back-spliced isoforms and some circular isoforms, in which the specific exons are expressed by a single gene site.10
DMD and DMBT1
There were 29 specific circRNA isoforms expressed in normal endometrial tissues and 14 expressed in EC tissues, among which the number of Dystrophin gene (DMD) isoforms decreased the most in EC tissues, from 29 in the normal endometrium to 14 in EC. However, the number of circRNA isoforms expressed by the Deleted In Malignant Brain Tumors 1 gene (DMBT1) was increased, from 32 in the normal endometrium to 50 in EC.10 Among the common hotspot genes expressed in normal and EC tissues, the circular transcriptional composition of these two genes changed significantly. DMD forms a specific circRNA in normal skeletal muscles. The specific circRNA formation may be the result of multi-exon skipping during DMD specific splicing.101 However, DMBT1 was elevated in biliary intraepithelial neoplasia, and its absence in the biliary tract was associated with poor survival in patients, suggesting that the expression of DMBT1 had an inhibitory effect on tumor growth.102 The special expression of DMD and DMBT1 in EC suggests that circRNA may be related to the development of EC.
DNAH14
DNAH14, which encodes the dynein heavy chain, is a unique hotspot gene in EC. DNAH14 produced 3 circular isoforms in normal endometrial tissue and 18 circular isoforms in EC tissue. With the increase of the number of DNAH14 isoforms, the expression of circular and linear transcripts in EC tissues were up-regulated. Chang et al found that DNAH14 is one of 21 passenger genes in EC, suggesting that DNAH14 abnormalities may interfere with cancer-related pathways.10,103
HSPG2 and RP11-255H23.4
Introns circRNA (HSPG2 and RP11255H23.4) were only expressed in normal endometrial tissue, but not in EC tissues. However, in endometrial tissues, the expression of miRNAs transcribed from their parent genes increased, indicating that these circRNAs can competitively bind to related miRNAs and play an important role in the occurrence and development of endometrial cancer. In the basement membrane, HSPG2 binds to growth factors to regulate the growth and regeneration of endothelial cells.10,16 This process is formed by its heparin sulfate glycosaminoglycan (HS-GAG) chain. And the decreased expression of HS-GAG was associated with EC progression.104
Although we do not know how each individual circRNAs expression contributes to the occurrence and development of tumors, it will be helpful and provide a basis for future studies on the functions and mechanisms of EC-related circRNAs.
Some CircRNAs That May Play a Role in EC as miRNA Sponges are Listed Below
Circ-ITCH
As microRNA (miRNA) sponges, circRNA protects target genes from miRNA-mediated mRNA cleavage, and is involved in the occurrence of various cancers such as liver cancer, gastric cancer and colorectal cancer. These circRNA-miRNA regulatory networks act on target genes involved in cell cycle regulation, signal transduction, epigenetic regulation or transcriptional regulation, and ultimately regulate the proliferation, differentiation, invasion and metastasis of cancer cells.15
Circ-ITCH is a circRNA produced by several exons of the itchy E3 ubiquitin protein ligase (ITCH) and tumor suppressor genes that act as sponges for specific miRNAs that target the ITCH parental transcript.49,105,106 Circ-ITCH can competitively bind to miRNA-17 and miRNA-224, a process that leads to differential expression of P21 and phosphatase and tensin homology (PTEN).107 P21 and PTEN are a cyclin-dependent kinase inhibitor or a well-known tumor suppressor. PTEN prevents angiogenesis in cancer tissue, inhibits cell division, proliferation, invasion and migration, and accelerates apoptosis.9 The mutation or deletion of PTEN inactivates its enzyme activity, thus losing its ability to inhibit cell proliferation. In addition, early studies have shown that various carcinogenic miRNAs promote the malignancy of tumors by neutralizing the expression of P21 or PTEN.108,109 Circ-ITCH is down-regulated in esophageal squamous cell carcinoma, colorectal cancer, hepatocellular carcinoma, and lung cancer by classical pathways. Circ-ITCH acts as a sponge for specific miRNAs, protecting the parent transcript ITCH and blocking downstream Wnt/β-catenin signaling, thereby preventing tumor progression.106,110–112 Studies have shown that circ-ITCH inhibits the aggressive biological behavior of BCa by stimulating the expression of target genes P21 and PTEN of miR-17 and miR-224 up-regulated by miR-17 and miR-224.104,107,113,114 Therefore, circ-ITCH inhibited BCa progression by eliminating the carcinogenic effects of miR-17 or miR-224 and forming the circ-ITCH/miR-17, miR-224/P21, PTEN axis. Therefore, we speculated that the differential expression of P21 and PTEN made the cells highly malignant and might develop into EC. In other words, P21 and PTEN may also perform similar regulation in EC through the sponge function of circRNAs.
Circ-ITCH targets tumor inhibition via novel circ-ITCH/miR-17, miR-224/P21, PTEN axis, which may provide a potential biomarker and target for EC treatment.
Hsa_Circ_0039569
Ye et al found that the expression of circRNAs in grade 3 EC tissues was different from that in matched non-tumor tissues. In women with grade 3 EC and adjacent non-cancerous endometrium tissue, a total of 62,167 unique circRNAs were significantly altered. Among them, 25,735 genes were significantly up-regulated and 36,432 genes were down-regulated. Among them, circRNAs such as hsa_circ_0039569, hsa_circ_0001523, hsa_circ_0001610, hsa_circ_0001400, hsa _circ_0007905 were up-regulated, while the circRNAs such as hsa_circ_0000437, hsa_circ_0009043, hsa_circ_0000471, and hsa_circ_0014606 were down-regulated.11 At the same time, these circRNAs were significantly differentially expressed in grade 3 EC and grade 1–2 EC.
The expression of hsa_circ_0039569 and hsa_circ_0001610 in grade 3 EC was more significant than that in grade1-2 EC. They have a higher level of expression in the grade 3 EC. Hsa_circ_0009043, hsa_circ_0000437 and hsa_circ_0001776 were significantly down-regulated in grade 3 EC and grade 1–2 EC tissues compared with adjacent non-malignant endometrium tissues. The expression levels of hsa_circ_0009043 and hsa_circ_0001776 in grade 3 non-cancer endometrium tissue were higher than that in grade 1 non-cancer endometrium tissue, while the expression levels of hsa_circ_0000437 were lower.11
The results of Ye et al also showed that the expression level of hsa_circ_0039569 was significantly correlated with tumor differentiation, but not with age, lymph node metastasis, tumor size, FIGO stage, or muscular invasion.
Most circRNAs that have a clear role in cancer act as miRNA sponges through the circRNA-miRNA axis.40,115–118 Studies have shown that interaction between hsa_circ_0039569 and hsa-miR-542-3p/hsa-let-7c-5p. And hsa-miR-542-3p and hsa-let-7c-5p were downregulated in the grade 3 EC. Therefore, hsa_circ_003956 was negatively correlated with hsa-miR-542-3p and hsa-let-7c-5p.11 Hsa_circ_0039569 can be used as an important predictor of level 3 EC.
Circ-ZNF91
ZNF91 belongs to a C2H2 zinc finger (ZNF) gene family, which has been greatly expanded in the primate lineage and is known to contain unusually rich targets of multiple miRNA families, including miR-23, miR-181 and miR-199.119 Circular ZNF91 had 24 binding sites with miR-23 and 7 binding sites with miR-199.120 Studies have shown that the expression of circular ZNF91 in EC is negatively correlated with the expression of miR-23B, miR-122A2 and miR-199.104 This suggests that circRNAs may act as miRNA sponges to inhibit the expression of miR-23B, miR-122A2 and miR-199.10 These miRNAs have been shown to be associated with a variety of human cancers, and their target genes and effects (promotion or inhibition) are different in different types of cancer. In prostate cancer, for example, miR-23B leads to decreased Sre expression, which slows tumor growth in nude mice. MiR-23B has also been shown to inhibit metastasis in colon cancer, and its targets include FZD7 or MAP3K1.121
Circ-8073
Cell cycle progression usually affects cell proliferation, and cell cycle disruption is considered to be a common cause of cell proliferation inhibition.122 Circ-8073 has been proved to be an important regulator of endometrial epithelial cell proliferation. As a miR-449a sponge, circ-8073 regulates endometrial epithelial cells (EECs) proliferation and cell cycle by regulating centrosomal protein of 55 (CEP55) expression. Circ-8073 gene knockout can induce EECs cell cycle arrest in G1/S phase. In addition, CEP55 promotes proliferation of glioma cells and reduces apoptosis through AKT/mTOR signaling pathway.123 Circ-8073 promotes EECs proliferation through the PI3K/AKT/mTOR pathway.124
CircPUM1
The expression level of circPUM1 in endometrial carcinoma tissues was significantly higher than that in normal tissues. Its upregulation can promote the proliferation, migration and invasion of endometrial cancer cells. CircPUM1 can bind to miR-136 and lead to the upregulation of NOTCH3, thus promoting the development of endometrial cancer.125 MiR-136 has been studied in a variety of cancers and has been identified as a tumor suppressor gene in a variety of adenocarcinomas such as colon, breast and lung cancers.126–128 Notch signaling affects many cellular processes, including being involved in cell fate decisions, maintaining undifferentiated states, inducing terminal differentiation, and other functions associated with cancer development.129 Therefore, circPUM1 promotes the development of EC by regulating miR-136/NOTCH3 axis.
Hsa_CircRNA_0001776
In EC tissues and cells, circ_0001776 and leucine-rich repeats and immunoglobulin-like domains 2 (LRIG2) expressions were down-regulated and miR-182 expressions were up-regulated.130 Low expression of circ_0001776 was associated with 5-year survival rate of EC patients. Up-regulation of circ_0001776 could inhibit cell proliferation and glycolysis, and promote cell apoptosis. In addition, circ_0001776 also regulated the expression of LRIG2 by acting as a sponge for miR-182, and CIRC_0001776 inhibited EC progression through miR-182/LRIG2 axis.11,131
Circ_0001776 has obvious inhibition on tumor growth in vivo. In other words, circ_0001776 inhibits the occurrence and development of EC through miR-182/LRIG2 axis, providing a potential target for the treatment of EC.131
Regulatory Pathways Involving QKI, CircRNA and ESRP2
The interactions between circRNAs and RNA-binding proteins have been proven to affect the progression of cancer.64 It has been reported that circRNA regulatory factor QKI protein level is positively correlated with 35 circRNAs,132 while epithelial splicing regulatory proteins 2 (ESRP2) level is negatively correlated with 20 circRNAs. These RBPs may act as major regulators of circRNAs. QKI is up-regulated in the epithelial to mesenchymal transition (EMT) process and promotes EMT by adjusting hundreds of variable splicing targets.132–134 It was found that relative QKI protein levels were positively correlated with EMT activators ZEB1 and ZEB2.85,135,136 ESRP2 levels, which play an important role in maintaining epithelial characteristics, were negatively correlated with QKI levels.137–139 ESRP2 regulates alternative splicing events associated with cellular epithelial phenotypes and plays a key role in EMT by regulating the isoforms of FGFR2, CD44, CTNND1, and ENAH.140,141
CircRNAs can act as miRNA sponges to regulate miRNA activity, while miRNAs play an important role in EMT.142 EMT is characterized by the transformation from polarized immobile epithelial cells to motional mesenchymal cells and is a powerful process of tumor metastasis, invasion and tumorigenesis.143,144 EMT is an important component of EC development and has prognostic significance.145 MiRNAs are important regulators of malignant transformation and metastasis. And many miRNAs are known to inhibit a variety of important cancer-related genes.146 It was found that the miR-200 family has abnormal expression in cancer and is involved in the initiation and progression of malignant transformation. The inhibitory effect of miR-200 members on metastasis is closely related to pathological EMT.147–150 Dou et al predicted miRNA binding sites in 35 circRNAs related to QKI level and found 36 potential binding sites of miRNA.85 It was also found that the activity of miRNA was negatively correlated with QKI expression.85
RNA-binding protein QKI was positively correlated with circRNAs, while QKI was negatively correlated with the activity of specific miRNAs, indicating a potential pattern that QKI, circRNAs and miRNAs form a regulatory feedback loop in EC.
The above is the summary of circRNAs that has an influence on the development of EC. At the same time, we also put these circRNAs and their functions in the table (Table 1). We found that most circRNAs function through the pathway of miRNA sponge, and altered miRNA expression plays a crucial role in the occurrence and development of endometrial cancer. Various miRNAs act as oncogenes or tumor suppressors and can regulate the occurrence and progression of tumors. Studies have found that after bortezomib treatment, the expression of miR-17-5p in endometrial cancer cells was decreased. It acts as an oncogene and acts in coordination with c-Myc, a oncogenic transcription factor that is often mutated or amplified in human cancers. A single microRNA may regulate a wide range of target genes, thus having a global impact on gene expression.151 Other studies have found that overexpression of miR-423 enhances the proliferation of endometrial cancer cells and increases their migration and invasion. MiR-423 has also been shown to play an important role in the tumorigenesis and development of endometrial cancer cells.152 MicroRNAs (miRNAs) involve in fine-tuning gene expression and releasing miRNAs that may lead to cascade cellular events that ultimately promote tumorigenesis.153–156 In addition, circRNAs can be released into the extracellular space and subsequently detected in the blood, plasma, serum and exosomes of gynecological cancer patients.157–160 Exosomes are small membranous vesicles of endocytic origin secreted by most cell types. They contain specific proteins, mRNAs and microRNAs that regulate the behavior of recipient cells and can be used as biomarkers for diagnosing human diseases.161 Studies have shown that circRNAs are enriched in exosomes compared to producing cells by RNA-seq analysis, and more than 1000 circRNAs have been identified in human serum exosomes. CircRNAs can bind to miRNAs, which are also abundant in exosomes.99 Serum exo-circRNA may distinguish cancer patients from healthy controls, suggesting its great translational potential as a biomarker in cancer diagnosis.
 |
Table 1 The Role of circRNAs in Endometrial Carcinoma |
Discussion and Prospects
In this review, we described the biogenesis, function and characteristics of circRNAs as well as their application in EC diagnosis and treatment. As we summarized in this review, circRNAs have an important role in the development of EC, and we summarizes the current circRNAs may have an impact on EC development. It is important to note that many studies have found that circRNAs abnormal expression in many tumor diseases, therefore they have great potential applications in the tumor treatment. Exogenous circRNAs may be ideal for diagnosis or therapeutic intervention in diseases due to its unique structure, high stability, and organ and tissue-specific expression patterns. Although more and more studies on circRNAs have provided us with a general understanding of the newest member of the RNA molecular world, the specific biogenesis and function of circRNAs are still unclear. At present, there are still many circRNAs to be studied.
CircRNAs abnormal expression of tumour disease has become the current hot research topic, but the circRNAs of abnormal expression of different diseases are different. With regard to the expression of circRNA in EC, few specific studies have carried out, and the sample size of the studies in this area is very small. At the same time, because of the complexity of the tumor pathogenesis, exact function of circRNAs is not clear in the EC. Therefore, the use of circRNAs as a biomarker for the diagnosis and prognosis of EC disease needs further study and is far from clinical application. The research of circRNAs in the field of disease is still in the preliminary stage, and there is still a lot of research to be done before it can be further developed.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Amant F, Moerman P, Neven P, et al. Endometrial cancer. Lancet. 2005;366(9484):491–505. doi:10.1016/s0140-6736(05)67063-8
2. Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi:10.3322/canjclin.55.1.10
3. Braun MM, Overbeek-Wager EA, Grumbo RJ. Diagnosis and Management of Endometrial Cancer. Am Fam Physician. 2016;93:468–474.
4. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi:10.1016/0090-8258(83)90111-7
5. Tornesello ML, Faraonio R, Buonaguro L, et al. The Role of microRNAs, Long Non-coding RNAs, and Circular RNAs in Cervical Cancer. Front Oncol. 2020;10:150. doi:10.3389/fonc.2020.00150
6. Huang J, Zhou Q, Li Y. Circular RNAs in gynecological disease: promising biomarkers and diagnostic targets. Biosci Rep. 2019;39. doi:10.1042/bsr20181641
7. Yang X, Mei J, Wang H, et al. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int. 2020;20:265. doi:10.1186/s12935-020-01367-9
8. Dong P, Xu D, Xiong Y, et al. The Expression, Functions and Mechanisms of Circular RNAs in Gynecological Cancers. Cancers. 2020;12. doi:10.3390/cancers12061472
9. Liu KS, Pan F, Mao XD, Liu C, Chen YJ. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am J Transl Res. 2019;11:1–15.
10. Chen BJ, Byrne FL, Takenaka K, et al. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2018;9(5):5786–5796. doi:10.18632/oncotarget.23534
11. Ye F, Tang QL, Ma F, et al. Analysis of the circular RNA transcriptome in the grade 3 endometrial cancer. Cancer Manag Res. 2019;11:6215–6227. doi:10.2147/cmar.S197343
12. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi:10.1038/nbt.2890
13. Ashwal-Fluss R, et al. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol Cell. 2014;56:55–66. doi:10.1016/j.molcel.2014.08.019
14. Vo JN, et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–881.e813. doi:10.1016/j.cell.2018.12.021
15. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi:10.1080/15476286.2015.1122162
16. Tran AM, et al. A New World of Biomarkers and Therapeutics for Female Reproductive System and Breast Cancers: circular RNAs. Front Cell Dev Biol. 2020;8:50. doi:10.3389/fcell.2020.00050
17. Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell. 2014;159:13–14. doi:10.1016/j.cell.2014.09.005
18. Starke S, et al. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015;10:103–111. doi:10.1016/j.celrep.2014.12.002
19. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna. 2015;21:172–179. doi:10.1261/rna.048272.114
20. Zhang XO, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi:10.1101/gr.202895.115
21. Chen -L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi:10.1038/s41580-020-0243-y
22. Rong D, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi:10.18632/oncotarget.19154
23. Wilusz JE. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9:e1478. doi:10.1002/wrna.1478
24. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi:10.1093/nar/gkr1009
25. Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14:978–984. doi:10.1080/15476286.2017.1317911
26. Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829–1842. doi:10.1261/rna.047126.114
27. Zhang Y, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi:10.1016/j.molcel.2013.08.017
28. Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–157. doi:10.1261/rna.035667.112
29. Czubak K, Sedehizadeh S, Kozlowski P, Wojciechowska M. An Overview of Circular RNAs and Their Implications in Myotonic Dystrophy. Int J Mol Sci. 2019;20. doi:10.3390/ijms20184385
30. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579. doi:10.1002/wrna.1294
31. Kristensen LS, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi:10.1038/s41576-019-0158-7
32. Ivanov A, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi:10.1016/j.celrep.2014.12.019
33. Du WW, et al. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. 2017;7:4183–4191. doi:10.7150/thno.21299
34. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi:10.1080/15476286.2015.1020271
35. Taylor K, et al. MBNL splicing activity depends on RNA binding site structural context. Nucleic Acids Res. 2018;46:9119–9133. doi:10.1093/nar/gky565
36. Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi:10.1371/journal.pbio.0020391
37. Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi:10.1101/gr.2855504
38. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi:10.1126/science.1064921
39. Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi:10.1038/nature09144
40. Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi:10.1038/nature11993
41. Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi:10.1016/j.cell.2011.09.028
42. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi:10.1371/journal.pone.0030733
43. Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi:10.1038/emboj.2011.359
44. Capel B, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi:10.1016/0092-8674(93)90279-y
45. Kefas B, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi:10.1158/0008-5472.Can-07-6639
46. Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi:10.1158/0008-5472.Can-08-2103
47. Bolisetty MT, Graveley BR. Circuitous route to transcription regulation. Mol Cell. 2013;51:705–706. doi:10.1016/j.molcel.2013.09.012
48. Li Z, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi:10.1038/nsmb.2959
49. Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928
50. Du WW, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–1412. doi:10.1093/eurheartj/ehw001
51. Du WW, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi:10.1093/nar/gkw027
52. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi:10.1126/science.7536344
53. Pamudurti NR, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21. doi:10.1016/j.molcel.2017.02.021
54. Legnini I, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22–37.e29. doi:10.1016/j.molcel.2017.02.017
55. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859:1245–1251. doi:10.1016/j.bbagrm.2016.07.009
56. AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A. 2014;111:14542–14547. doi:10.1073/pnas.1402814111
57. Flores R, et al. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 2011;8:200–206. doi:10.4161/rna.8.2.14238
58. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi:10.1038/323558a0
59. Zhang M, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475. doi:10.1038/s41467-018-06862-2
60. Ye F, et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol Ther Nucleic Acids. 2019;18:88–98. doi:10.1016/j.omtn.2019.07.023
61. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi:10.3390/ijms15069331
62. Huang C, Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6:61–64. doi:10.1080/21541264.2015.1071301
63. Lei K, et al. The mechanism and function of circular RNAs in human diseases. Exp Cell Res. 2018;368:147–158. doi:10.1016/j.yexcr.2018.05.002
64. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi:10.7150/thno.42174
65. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi:10.15252/embj.2018100836
66. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi:10.1371/journal.pgen.1003777
67. Maass PG, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 2017;95:1179–1189. doi:10.1007/s00109-017-1582-9
68. Perriman R, Ares M. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. Rna. 1998;4:1047–1054. doi:10.1017/s135583829898061x
69. Haddad G, Lorenzen JM. Biogenesis and Function of Circular RNAs in Health and in Disease. Front Pharmacol. 2019;10:428. doi:10.3389/fphar.2019.00428
70. Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi:10.1101/gad.314856.118
71. Cai H, et al. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol. 2019;234:11391–11400. doi:10.1002/jcp.27796
72. Gao YL, et al. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8:86625–86633. doi:10.18632/oncotarget.21257
73. Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604.
74. Tian JDC, Liang L. Involvement of circular RNA SMARCA5/microRNA-620 axis in the regulation of cervical cancer cell proliferation, invasion and migration. Eur Rev Med Pharmacol Sci. 2018;22:8589–8598. doi:10.26355/eurrev_201812_16622
75. Zhang J, Zhao X, Zhang J, Zheng X, Circular LF. RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem Biophys Res Commun. 2018;501:428–433. doi:10.1016/j.bbrc.2018.05.006
76. Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235:718–724. doi:10.1002/jcp.29009
77. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:3713–3718. doi:10.26355/eurrev_201806_15250
78. Hill HA, et al. Racial differences in endometrial cancer survival: the black/white cancer survival study. Obstet Gynecol. 1996;88:919–926. doi:10.1016/s0029-7844(96)00341-9
79. Madison T, Schottenfeld D, James SA, Schwartz AG, Gruber SB. Endometrial cancer: socioeconomic status and racial/ethnic differences in stage at diagnosis, treatment, and survival. Am J Public Health. 2004;94:2104–2111. doi:10.2105/ajph.94.12.2104
80. Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262
81. Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi:10.1016/s0140-6736(15)00130-0
82. Zaino RJ, Kurman RJ, Diana KL, Morrow CP. The utility of the revised International Federation of Gynecology and Obstetrics histologic grading of endometrial adenocarcinoma using a defined nuclear grading system. A Gynecologic Oncology Group study. Cancer. 2015;75:81–86.
83. Luoto R, Raitanen J, Pukkala E, Anttila A. Effect of hysterectomy on incidence trends of endometrial and cervical cancer in Finland 1953-2010. Br J Cancer. 2004;90:1756–1759. doi:10.1038/sj.bjc.6601763
84. Schouten LJ, Goldbohm RA, van den Brandt PA. Anthropometry, physical activity, and endometrial cancer risk: results from the Netherlands Cohort Study. J Natl Cancer Inst. 2004;96:1635–1638. doi:10.1093/jnci/djh291
85. Dou Y, et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell. 2020;180:729–748.e726. doi:10.1016/j.cell.2020.01.026
86. Vausort M, et al. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J Am Coll Cardiol. 2016;68:1247–1248. doi:10.1016/j.jacc.2016.06.040
87. Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. 2017;17:33–36. doi:10.1016/j.ijcha.2017.11.001
88. Chen B, et al. circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics. 2018;8:4003–4015. doi:10.7150/thno.24106
89. Chen X, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–671. doi:10.1080/15548627.2019.1634945
90. Liu CX, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177:865–880.e821. doi:10.1016/j.cell.2019.03.046
91. Carrara M, Fuschi P, Ivan C, Martelli F. Circular RNAs: methodological challenges and perspectives in cardiovascular diseases. J Cell Mol Med. 2018;22:5176–5187. doi:10.1111/jcmm.13789
92. Chen S, et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell. 2019;176:831–843.e822. doi:10.1016/j.cell.2019.01.025
93. Dragomir M, Calin GA. Circular RNAs in Cancer - Lessons Learned From microRNAs. Front Oncol. 2018;8:179. doi:10.3389/fonc.2018.00179
94. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi:10.1038/onc.2017.361
95. Yang Y, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi:10.1093/jnci/djx166
96. Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–1064. doi:10.1002/cbin.10826
97. Zheng Q, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi:10.1038/ncomms11215
98. Bachmayr-Heyda A, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi:10.1038/srep08057
99. Li Y, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi:10.1038/cr.2015.82
100. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10:e0141214. doi:10.1371/journal.pone.0141214
101. Suzuki H, Aoki Y, Kameyama T, et al. Endogenous Multiple Exon Skipping and Back-Splicing at the DMD Mutation Hotspot. Int J Mol Sci. 2016;17(10):1722. doi:10.3390/ijms17101722
102. Goeppert B, Roessler S, Becker N, et al. DMBT1 expression in biliary carcinogenesis with correlation of clinicopathological data. Histopathology. 2017;70(7):1064–1071. doi:10.1111/his.13175
103. Chang YS, Huang HD, Yeh KT, Chang JG. Identification of novel mutations in endometrial cancer patients by whole-exome sequencing. Int J Oncol. 2017;50:1778–1784. doi:10.3892/ijo.2017.3919
104. Kodama J, Kusumoto T. Loss of basement membrane heparan sulfate expression is associated with tumor progression in endometrial cancer.. Eur J Gynaecol Oncol. 2005;26(4):403–406.
105. Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14(1):10–21. doi:10.1016/j.ccr.2008.06.001
106. Huang G, Zhu H, Shi Y, et al. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. 2015;10(6):e0131225. doi:10.1371/journal.pone.0131225
107. Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19. doi:10.1186/s12943-018-0771-7
108. Gu J, Wang D, Zhang J, et al. GFRα2 prompts cell growth and chemoresistance through down-regulating tumor suppressor gene PTEN via Mir-17-5p in pancreatic cancer. Cancer Lett. 2016;380(2):434–441. doi:10.1016/j.canlet.2016.06.016
109. Ye J, Yao Y, Song Q, et al. Up-regulation of miR-95-3p in hepatocellular carcinoma promotes tumorigenesis by targeting p21 expression. Sci Rep. 2016;6(1):34034. doi:10.1038/srep34034
110. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi:10.18632/oncotarget.3469
111. Wan L, Zhang L, Fan K, et al. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/β-Catenin Pathway.. Biomed Res Int. 2016;2016:1579490. doi:10.1155/2016/1579490
112. Guo W, Zhang J, Zhang D, et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8(29):48169–48177. doi:10.18632/oncotarget.18327
113. Tang K, Wang C, Chen Z, Xu H, Ye Z. Clinicopathologic and prognostic significance of p21 (Cip1/Waf1) expression in bladder cancer.. Int J Clin Exp Pathol. 2005;26(5):4999–5007.
114. Cazier JB, Rao SR, McLean CM, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014;5(1):3756. doi:10.1038/ncomms4756
115. Anderson DM, Anderson K, Chang C-L, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi:10.1016/j.cell.2015.01.009
116. Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi:10.18632/oncotarget.8589
117. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919–3931. doi:10.1038/onc.2015.460
118. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi:10.1038/srep30919
119. Schnall-Levin M, et al. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21:1395–1403. doi:10.1101/gr.121210.111
120. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi:10.1186/s13059-014-0409-z
121. Zhang H, et al. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. doi:10.1038/ncomms1555
122. Greenbaum LE. Cell cycle regulation and hepatocarcinogenesis. Cancer Biol Ther. 2004;3:1200–1207. doi:10.4161/cbt.3.12.1392
123. Wang G, et al. Centrosomal Protein of 55 Regulates Glucose Metabolism, Proliferation and Apoptosis of Glioma Cells via the Akt/mTOR Signaling Pathway. J Cancer. 2016;7:1431–1440. doi:10.7150/jca.15497
124. Liu X, et al. Circ-8073 regulates CEP55 by sponging miR-449a to promote caprine endometrial epithelial cells proliferation via the PI3K/AKT/mTOR pathway. Biochim Biophys Acta Mol Cell Res. 2018;1865:1130–1147. doi:10.1016/j.bbamcr.2018.05.011
125. Zong ZH, Liu Y, Chen S, Circ_PUM1 ZY. promotes the development of endometrial cancer by targeting the miR-136/NOTCH3 pathway. J Cell Mol Med. 2020;24:4127–4135. doi:10.1111/jcmm.15069
126. Yan M, et al. miR-136 suppresses tumor invasion and metastasis by targeting RASAL2 in triple-negative breast cancer. Oncol Rep. 2016;36:65–71. doi:10.3892/or.2016.4767
127. Ren H, Qi Y, Yin X, Gao J. miR-136 targets MIEN1 and involves the metastasis of colon cancer by suppressing epithelial-to-mesenchymal transition. Onco Targets Ther. 2018;11:67–74. doi:10.2147/ott.S113359
128. Shen S, et al. Upregulation of miR-136 in human non-small cell lung cancer cells promotes Erk1/2 activation by targeting PPP2R2A. Tumour Biol. 2014;35:631–640. doi:10.1007/s13277-013-1087-2
129. Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi:10.1038/nrc1186
130. Devor EJ, et al. Cullin-5, a ubiquitin ligase scaffold protein, is significantly underexpressed in endometrial adenocarcinomas and is a target of miR-182. Oncol Rep. 2016;35:2461–2465. doi:10.3892/or.2016.4605
131. Jia Y, Liu M, Wang S. CircRNA hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis in endometrial cancer via downregulating LRIG2 by sponging miR-182. Cancer Cell Int. 2020;20:412. doi:10.1186/s12935-020-01437-y
132. Conn SJ, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi:10.1016/j.cell.2015.02.014
133. Nieto MA, Huang RY, Jackson RA, Thiery JPEMT. (2016). Cell. 2016;166:21–45. doi:10.1016/j.cell.2016.06.028
134. Pillman KA, et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018;37. doi:10.15252/embj.201899016
135. Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–529. doi:10.1038/ncb3513
136. Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi:10.1080/15384101.2015.1006048
137. Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol. 2012;22:417–427. doi:10.1016/j.semcancer.2012.04.003
138. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi:10.1016/j.molcel.2009.01.025
139. Mizutani A, Koinuma D, Seimiya H, Miyazono K. The Arkadia-ESRP2 axis suppresses tumor progression: analyses in clear-cell renal cell carcinoma. Oncogene. 2016;35:3514–3523. doi:10.1038/onc.2015.412
140. Ishii H, et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014;289:27386–27399. doi:10.1074/jbc.M114.589432
141. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi:10.1038/nrm3758
142. Zaravinos A. The Regulatory Role of MicroRNAs in EMT and Cancer. J Oncol. 2015;2015:865816. doi:10.1155/2015/865816
143. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi:10.1007/s10555-008-9169-0
144. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi:10.1172/jci39104
145. Tanaka Y, et al. Prognostic impact of EMT (epithelial-mesenchymal-transition)-related protein expression in endometrial cancer. Cancer Biol Ther. 2013;14:13–19. doi:10.4161/cbt.22625
146. Zhang B, Pan X, Cobb GP, Anderson T. A. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi:10.1016/j.ydbio.2006.08.028
147. Adam L, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi:10.1158/1078-0432.Ccr-08-2245
148. Wiklund ED, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–1334. doi:10.1002/ijc.25461
149. Koutsaki M, Spandidos DA, Zaravinos A. Epithelial-mesenchymal transition-associated miRNAs in ovarian carcinoma, with highlight on the miR-200 family: prognostic value and prospective role in ovarian cancer therapeutics. Cancer Lett. 2014;351:173–181. doi:10.1016/j.canlet.2014.05.022
150. Díaz-López A, Moreno-Bueno G, Cano A. Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag Res. 2014;6:205–216. doi:10.2147/cmar.S38156
151. Shen Y, et al. Bortezomib induces apoptosis of endometrial cancer cells through microRNA-17-5p by targeting p21. Cell Biol Int. 2013;37:1114–1121. doi:10.1002/cbin.10139
152. Li J, Sun H, Liu T, Kong J. MicroRNA-423 promotes proliferation, migration and invasion and induces chemoresistance of endometrial cancer cells. Exp Ther Med. 2018;16:4213–4224. doi:10.3892/etm.2018.6710
153. Chou YT, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70:8822–8831. doi:10.1158/0008-5472.Can-10-0638
154. Gottardo F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi:10.1016/j.urolonc.2007.01.019
155. Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi:10.1053/j.gastro.2006.02.057
156. Zhang N, Li X, Wu C, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32:5078–5088. doi:10.1038/onc.2012.526
157. Gao Y, Zhang C, Liu Y, Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends. 2019;13:204–211. doi:10.5582/bst.2019.01021
158. Wang J, Wu A, Yang B, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in ovarian cancer. Gene. 2020;724:144150. doi:10.1016/j.gene.2019.144150
159. Guan X, Zong ZH, Liu Y, et al. circPUM1 Promotes Tumorigenesis and Progression of Ovarian Cancer by Sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892. doi:10.1016/j.omtn.2019.09.032
160. Wang W, Wang J, Zhang X, Liu G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:7451–7457. doi:10.2147/ott.S220700
161. Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi:10.1016/j.bcp.2011.12.037
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.