Back to Journals » Cancer Management and Research » Volume 13
Circular RNA_0000629 Suppresses Bladder Cancer Progression Mediating MicroRNA-1290/CDC73
Authors Wang J, Luo J, Wu X, Gao Z
Received 18 November 2020
Accepted for publication 15 February 2021
Published 22 March 2021 Volume 2021:13 Pages 2701—2715
DOI https://doi.org/10.2147/CMAR.S292863
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Jiansong Wang, Jianjun Luo, Xuecheng Wu, Zhiyong Gao
Department of Urology, Hunan Provincial People’s Hospital, The First Affiliated Hospital of Hunan Normal University, Changsha, 410005, Hunan, People’s Republic of China
Correspondence: Zhiyong Gao
Department of Urology, Hunan Provincial People’s Hospital, The First Affiliated Hospital of Hunan Normal University, No. 61, Jie Fang Xi Road, Changsha, 410005, Hunan, People’s Republic of China
Tel/Fax +86 0731-83929411
Email [email protected]
Background: Recent studies showed circular RNAs (circRNAs) played regulatory roles in bladder cancer (BC). However, the relevance of circ_0000629, a newly identified circRNA, has not been determined yet. We aimed to characterize the function of circ_0000629 in BC and the relevant mechanism.
Methods: First, we downloaded circRNA-related microarrays GSE147985 and GSE92675 from the GEO database, followed by a validation in our clinically obtained samples. We then overexpressed circ_0000629 in T24 and SW780 cells and evaluated the effects of circ_0000629 on BC cell proliferatory, apoptotic, and metastatic abilities. We further detected the subcellular localization of circ_0000629 in T24 and SW780 cells by the fractionation and export assay and FISH experiments. Integrated microarray analyses and bioinformatics website prediction were utilized to screen out the downstream microRNA (miRNA)/mRNA. The effects of miR-1290 and CDC73 on BC cell growth and metastasis was verified by functional rescue experiments. In addition, mice xenografts were built to measure the effect of circ_0000629 on tumor growth in vivo.
Results: Circ_0000629 and CDC73 were reduced, and miR-1290 was significantly overexpressed in BC tissues and cells. Moreover, circ_0000629 significantly inhibited the development and metastasis of BC cells, but further overexpression of miR-1290 or knockdown of CDC73 attenuated the inhibitory effect of circ_0000629 on BC cells. Circ_0000629 localized in the cytoplasm and regulated CDC73 expression by sponging miR-1290. Further, overexpressed circ_0000629 reduced the BC tumor growth in vivo.
Conclusion: Circ_0000629 promotes the expression of CDC73 by competitively binding to miR-1290, thereby inhibiting the growth and metastasis of BC cells.
Keywords: bladder cancer, circular RNA_0000629, microRNA-1290, CDC73, epigenetics
Introduction
Bladder cancer (BC) is the ninth most frequent malignancy worldwide, with an estimated 430,000 new diagnoses in 2012, and more than 60% of all BC cases and half of all the 165,000 deaths occur in the developing countries.1 Tobacco smoke in addition to occupational and environmental carcinogens feed into the risk factors of BC.2 The most frequently-seen clinical presentation for BC is asymptomatic hematuria, which should prompt assessment with cystoscopy, renal function examination, and upper urinary tract imaging in adults and in those with irritative voiding symptoms, risk factors for BC, or gross hematuria at any age.3 Moreover, the high rates of recurrence and substantial risk of development into higher-grade tumors mandate other therapies with intravesical agents.4
The genetic modulation (genetic mutation and epigenetic modifications) is found to be responsible for the pathogenesis of BC, among which non-coding RNAs have been identified in the mediation of gene expression and disease progressions.5 Circular RNAs (circRNAs), first discovered in plant viroids in 1976, are a family of non-coding RNAs which possess unique covalent loop structures that generally accumulate in the cytoplasm.6,7 In particular, the significance of circRNAs has been underscored in the process of tumorigenesis, invasion, and metastasis, which makes them potential diagnostic and therapeutic biomarkers of BC.8 CircRNAs have been revealed to function as microRNA (miRNA) sponges, regulators of transcription, as well as modifiers of gene expression.9 For instance, circ-FOXO3 positively modulated TGFBR2 expression by sponging miR-9-5p in BC cells.10 In the present study, we resorted to Gene Expression Omnibus (GEO) databases for determining the targets. Circ_0000629, a novel circRNA that has been rarely explored before, was found to be the only circRNAs intersected by both GSE147985 and GSE92675 dataset. Subsequent prediction of miRNA target of circ_0000629 and miRNA upregulated in BC helped us determine that miR-1290 might be a downstream biomolecule of circ_0000629. Serum exosomal miR-1290 has been found to be significantly overexpressed in lung adenocarcinoma patients relative to healthy controls, and its expression was closely linked to tumor stage, lymph node metastasis as well as distant metastasis.11 Moreover, another circRNA hsa_circ_0008309 has been suggested to have the potency to interact with miR-1290 and to regulate the downstream gene expression in oral squamous cell carcinoma.12 Therefore, we postulated that circ_0000629 could bind to miR-1290 to mediate the expression of a downstream molecule, thus involving in the progression of BC. The proposed circ_0000629 function might add new evidence for its role as a biomarker of BC.
Materials and Methods
Data Mining
From GEO database (https://www.ncbi.nlm.nih.gov/geo/), clinical samples of BC were obtained. First, the gene expression datasets of BC and adjacent tissues were processed in R language to obtain expression profiles. Subsequently, the target miRNAs of circ_0000629 were predicted using the CircBank website (http://www.circbank.cn/). Alternatively, the TargetScan (http://www.targetscan.org/vert_72/) and miRDB (http://mirdb.org/) websites predicted the target mRNAs of miR-1290.
Clinical Samples
A total of 57 patients who underwent radical cystectomy at Hunan Provincial People’s Hospital from March 2013 to December 2018 was collected. Participants were selected based on inclusion criteria: patients were 18 years of age or older; had a verified histopathological diagnosis (myxomatous disease, pT2-T4, N0-N3, M0), and had complete follow-up data. Exclusion criteria: BC patients had distant metastases; patients had non-muscle invasive BC; BC patients received neoadjuvant chemotherapy prior to surgery. All patients included received laparoscopic or open radical cystectomy and standard bilateral lymphadenectomy. Tumor staging was conducted based on tumor node metastasis staging system proposed by Union Internationale Contre le Cancer (International Union Against Cancer), and tumor grading on the basis of the 2004 World Health Organization classification system. Tumor sections were reviewed by two urological pathologists to confirm the diagnosis. This study was approved by the Research and Ethical Committee of Hunan Provincial People’s Hospital and conducted in accordance with the Declaration of Helsinki. Participants signed informed consents prior to sample collection.
Real-Time Quantitative PCR (RT-qPCR)
Total RNA was isolated using Trizol (Ambion, Austin, TX, USA) as per the manufacturer’s instructions. RNA was reverted into complementary DNA (cDNA) using the Promega M-MLV kit. qPCR was carried out with the help of SYBR Select Master Mix (Applied Biosystems, Inc., Foster City, CA, USA) on the Applied Biosystems 7500 Fast platform (Applied Biosystems). After normalization with reference to GAPDH expression, the relative expression was calculated as 2−ΔΔCT method. Primer sequences are listed in Table 1.
 |
Table 1 Primers |
Cells
The normal human bladder uroepithelial cell line (SV-HUC-1) and four BC cell lines (5637, J82, T24, and SW780) were acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). SV-HUC-1, T24, and SW780 cells were grown in DMEM (Gibco, Carlsbad, CA, USA). 5637 and J82 were in Roswell Park Memorial Institute-1640 medium (Gibco). All media contained 1% penicillin/streptomycin and 10% FBS (Gibco). The culture environment was humidified air enriched with 5% CO2 at 37°C.
Subsequently, T24 and SW780 cells in good growth condition were infected using the lentiviral vector overexpressing circ_0000629 and empty vector alone or with miR-1290 mimic and short hairpin RNA (shRNA) targeting CDC73. Successful overexpression was confirmed 48 h after infection using RT-qPCR by detecting the expression of circ_0000629 in the cells.
Cell Counting Kit-8 (CCK-8) Assay
Cell plated into 96-well plates were maintained overnight at 37°C. At the 0, 12th, 24th, 48th, and 72nd hours post-transfection, cells were treated with CCK-8 (10 μL/well) and incubated for 1 h. Subsequently, the optical density (OD) value was read at 450 nm.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Staining
A total of 1 × 104 cells/well of T24 and SW780 cells were plated in 96-well plates. After 12 h, cells were stained for 2 h with EdU reagent included in the Cell-Light EdU Apollo567 In Vitro Kit (RiboBio, Guangzhou, Guangdong, China). Three sets of photographs (400× magnification) of each group of well-stained cells were taken randomly under a fluorescence microscope (Nikon, Tokyo, Japan). The ratio of EdU-positive cells (Apollo567-stained cells/Hoechst-stained cells) was then calculated.
Flow Cytometry
Apoptosis was detected using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection Kit (Beyotime, Shanghai, China). Briefly, cells were collected with trypsin/ethylenediaminetetraacetic acid and resuspended in binding buffer. Next, cells were cultured with 5 μL Annexin V-FITC at 4°C for 15 min and with 5 µL PI at 4°C for 5 min. After incubation, apoptotic cells were quantified by a flow cytometer (BD Biosciences, San Jose, CA, USA), and data analysis was implemented using BD AccuriTM C6 software.
Caspase-3/7 Activity Assay
To analyze the caspase-3 and caspase-7 activity in T24 and SW780 cells, a caspase-Glo 3/7 assay (Promega, Madison, WI, USA) was used, and luminescence was measured. Therefore, 9 × 105 T24 and SW780 cells were plated in 300 μL growth medium in 24-well plates and incubated overnight. After 24 h, 80 μL cell suspension was mixed with 80 μL caspase-3/7 reagent and seeded in a 96-well plate, and luminescence was measured after 2 h of incubation (Orion II microplate photometer).
Western Blot
Total protein was extracted from the cells. Approximately 25 μg sample was processed by 12% polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes. The membranes were placed in a blocking solution and slowly shaken for 2 h at 37°C. After blocking, the membranes were mixed with primary antibodies to PUMA (ab33906, 1:1000, Abcam, Cambridge, UK), Bax (#33-6400, 1:500, Invitrogen Inc., Carlsbad, CA, USA), Bcl-2 (#MA5-11757, 1:1000, Invitrogen), E-cadherin (GTX100443, 1:500, GeneTex, Inc., Alton Pkwy Irvine, CA, USA), Vimentin (GTX100619, 1:1000, GeneTex), Snai1 (#14-9859-82, 1:500, Invitrogen), N-cadherin (ab70611, 1:2000, Abcam), CDC73 (#PA5-17159, 1:1000, Invitrogen) and GAPDH (ab8245, 1:2000, Abcam) overnight at 4°C. The membranes were then incubated with the secondary antibody to IgG labelled with horseradish peroxidase (ab6721, 1:1000, Abcam) for 2 h at 37°C. Protein signals were visualized using enhanced chemiluminescence. The protein levels were analyzed using ImageJ software. The relative expression of the target protein was obtained by calculating the ratio of optical density (OD) value of the target protein to that of an internal reference.
Transwell Assay
Cell migration and invasion were evaluated using the Transwell chamber assays. For the cell migration assay, cells were plated at 2 × 104 cells per well in the apical chamber (8.0 μm pore size polycarbonate membrane, BD Biosciences) and filled with serum-free medium. The basolateral chamber was supplemented with DMEM containing 10% FBS. For the invasion assay, cell invasion was determined by using a 24-well Matrigel-coated invasion chamber (BD Biosciences) with a pore size of 8 μm. The rest procedure was the same as the migration assay. Cells were incubated at 37°C in a 5% CO2 incubator for 12 h (migration assay) or 24 h (invasion assay). Afterwards, non-migrated or non-invaded cells were removed from the surface of the membrane by wiping with a cotton swab. Cells on the underside of the membrane were fixed in 4% paraformaldehyde for 15 min and stained with 1% crystal violet solution for 5 min. Adherent cells in five fields were randomly captured with a microscope (Olympus, Tokyo, Japan). Migrated/invaded cells were calculated using Image-Pro Plus software.
Fractionation and Export Assay
Cytoplasmic and nuclear RNA was isolated from T24 and SW780 cells using the Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA). The rate of the indicated RNA molecule expression in the nucleus to that in the cytoplasm was then determined by RT-qPCR assay. U6 was selected as the nucleus control and GAPDH as the cytoplasmic control.
FISH
Briefly, T24 and SW780 cells were seeded on cell slides, fixed with 4% paraformaldehyde, treated with 0.5% Triton and prehybridized. Overnight hybridization was then performed with probes (10 mM). The assays were conducted using RNA FISH kit (Guangzhou RiboBio Co., Ltd., Guangzhou, Guangdong, China) according to the manufacturer’s instructions. The Cy3-labeled circ_0000629 probe was synthesized and provided by Sangon Biotech (Shanghai, China). Images were taken with a confocal microscope (Carl Zeiss, Oberkochen, Germany).
Dual-Luciferase Reporter Assay
For the circ_0000629 and CDC73 luciferase reporter assays, we synthesized circ_0000629 and CDC73 fragments containing predicted miR-1290 binding sites (circ_0000629-wt and CDC73-wt) or mutant fragments (circ_0000629-mt and CDC73-mt). The synthesized fragments were then inserted downstream of the luciferase reporter gene in the pmirGLO vector (Promega, Madison, WI, USA). For miRNA target gene luciferase reporter assays, the synthesized CDC73 bind sites to miR-1290 or mutation site were inserted into the pmirGLO vector. The constructed vectors were transfected into cells using either miR-1290 mimic or mimic NC with the help of Lipofectamine 3000 following the manufacturer’s instructions. Renilla and firefly luciferase activities were assessed using a dual-luciferase reporter system (Promega). Renilla luciferase activity served as a normalized control to calculate relative luciferase activity.
Mouse Xenograft Models
Experimental procedures were approved by the Institutional Committee on the Care and Use of Animals of Hunan Provincial People’s Hospital. All animals received humane care according to the National Institutes of Health (USA) guidelines. Specific-pathogen-free BALB/c-nu thymus-free nude mice (4–5 weeks old, male, Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were used and divided into four groups of six animals each. Stable cell lines T24 and SW780 (5 × 106) overexpressing circ_0000629 and Vector NC were inoculated subcutaneously into the axilla of mice. Tumor length and width were measured once every 5 days, and the tumor volume was calculated using the formula: volume (mm3) = width2 × length/2. After 35 days, the mice were euthanized, and the tumors were removed to perform further experiments.
Immunohistochemistry
Tumor tissues were fixed with 4% paraformaldehyde, paraffin-embedded, and sectioned. Sections were recovered in Dulbecco’s phosphate buffered saline buffer for antigen recovery, blocked with a drop of goat serum at room temperature, followed by the incubation with the primary antibody against KI67 (1:200, ab15580, Abcam) overnight at 4°C. The next day, the color development reaction was performed with diaminobenzidine chromogenic solution (Dalian Meilun Biotechnology Co., Ltd., Dalian, Liaoning, China), and then the nuclei were counter-stained with hematoxylin (Wuxi Jiangyuan Industrial Technology & Trade Ltd., Wuxi, Jiangsu, China). The slides were then dehydrated, dried at room temperature, and sealed with neutral resin.
Data Analysis
GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA, USA) was utilized for the analysis of data, which were presented as mean ± SD. Results were analyzed using paired t-test, or one-way or two-way analysis of variance (ANOVA) test. All experiments were repeated at least three times. A p value of less than 0.05 was deemed as statistically significant.
Results
Circ_0000629 is Reduced in BC Tissues and Cell Lines
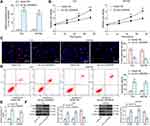
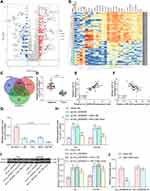
We first downloaded circRNA gene expression microarrays for BC from the GEO database GSE147985 which contains cancerous and adjacent tissues from four BC patients and GSE92675 which also contains cancerous and adjacent tissues from four BC patients. We screened out 65 and 461 differentially expressed circRNAs, respectively (Figure 1A and B) by setting Log FoldChange > 1, adj p value < 0.05 as the thresholds after data collection as well as normalization. Only circ_0000629 was observed in the intersection (Figure 1C). After that, the expression of this circRNA was evaluated in cancerous and adjacent tissues from 57 BC patients by RT-qPCR. Circ_0000629 was significantly poorly expressed in BC tissues (Figure 1D). We further examined circ_0000629 expression in normal bladder uroepithelial cells and BC cell lines. Circ_0000629 was revealed to be significantly reduced in the BC cell lines (Figure 1E).
Overexpression of Circ_0000629 Inhibits the Growth of BC Cells in vitro
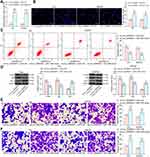
To further determine the role of circ_0000629 in the growth of BC cells, we transfected the overexpression plasmids of circ_0000629 into T24 and SW780 cells and confirmed successful transfections by RT-qPCR (Figure 2A). Subsequently, we examined cell activity using the CCK-8 kit, and we found that the cell activity of T24 and SW780 cells was significantly inhibited following overexpression of circ_0000629 (Figure 2B). The results of EdU staining showed that circ_0000629 significantly suppressed the number of EdU-positive cells in T24 and SW780 cells (Figure 2C). Moreover, we further used flow cytometry to detect the percentage of apoptosis. After overexpression of circ_0000629, flow cytometry revealed that the percentage of apoptosis in T24 and SW780 cells was significantly increased (Figure 2D) and the activity of caspase-3/7 in cells also significantly increased (Figure 2E). Subsequently, we examined the expression of the apoptosis-related proteins PUMA, Bax, and Bcl-2 using Western blot. The expression of PUMA and Bax was significantly increased and the expression of Bcl-2 was significantly decreased after overexpression of circ_0000629 in T24 and SW780 cells (Figure 2F).
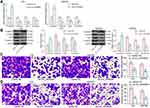
Overexpression of CircRNA_0000629 Inhibits Aggressiveness of BC Cells in vitro
Our aforementioned results showed that circ_0000629 overexpression significantly inhibited the growth of BC cells in vitro, whereas the effect on the migration and invasion of BC cells in vitro was unknown. Thus, we first detected the expression patterns of epithelial-mesenchymal transition (EMT)-related factors in T24 and SW780 cells using RT-qPCR and Western blot. E-cadherin was remarkably increased after overexpression of circ_0000629, while the expression of Vimentin, Snail, and N-cadherin were significantly suppressed (Figure 3A and B). Subsequently, we further used Transwell assays to detect changes in cell migration and invasion capacities. Overexpression of circ_0000629 significantly reduced the number of T24 and SW780 cells migrated or invaded into the Transwell basolateral chamber (Figure 3C and D). The above findings demonstrated that circ_0000629 can inhibit the growth and aggressiveness of BC cells in vitro.
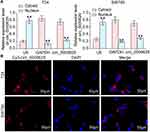
CircRNA_0000629 is Localized in the Cytoplasm
To further determine the regulatory role of circ_0000629 on BC cell growth and aggressiveness, we first detected the subcellular localization of circ_0000629 in BC cells using fractionation and export assays. Circ_0000629 was principally distributed in the cytoplasm (Figure 4A). Moreover, we further employed FISH experiments to confirm the subcellular localization of circ_0000629 in T24 and SW780 cells. We found that in T24 and SW780 cells, Cy3-tagged circ_0000629 was predominantly distributed around the blue nuclei stained by DAPI, ie circ_0000629 was predominantly distributed in the cytoplasm (Figure 4B).
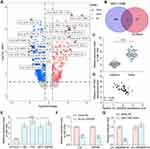
CircRNA_0000629 Interacts with miR-1290
To further determine the downstream regulatory mechanism of circ_0000629, we first downloaded the BC miRNA expression microarray GSE113486 from the GEO database, which contains 392 BC patients and 100 normal bladder tissues, and we screened a total of 418 differentially expressed miRNAs (Figure 5A). The target miRNAs of circ_0000629 were predicted through the circbank website and intersected with the miRNAs upregulated in the GSE113486 microarrays. miR-1290 was revealed to be in the intersection (Figure 5B). We then detected the miR-1290 expression in cancerous and adjacent tissues of 57 BC patients by RT-qPCR. miR-1290 was increased in cancerous tissues (Figure 5C) and inversely correlated with circ_0000629 expression (Figure 5D). Moreover, we further examined the miR-1290 expression in bladder uroepithelial cells and BC cells and observed that miR-1290 expression was remarkably increased in BC cells (Figure 5E). In addition, circ_0000629 upregulation significantly decreased the expression of miR-1290 in T24 and SW780 (Figure 5F). To further clarify the mechanism of regulation of miR-1290 by circ_0000629, we used dual-luciferase experiments. The luciferase activity in 293T cells delivered with miR-1290 mimic and circ_0000629-wt was much lower than that in cells transfected with mimic NC or circ_0000629-mt (Figure 5G).
miR-1290 Mimic Potentiates the Growth and Aggressiveness of BC Cells in vitro
To determine the role of miR-1290 in the growth of BC cells, we further transfected miR-1290 mimic in T24 and SW780 cells stably overexpressing circ_0000629 and verified successful transfection by RT-qPCR (Figure 6A). We noted that the growth activity of T24 and SW780 cells was significantly promoted after overexpression of miR-1290 (Figure 6B). By contrast, increased apoptosis due to oe-circ_0000629 was partially reversed by miR-1290 mimic (Figure 6C). Moreover, the protein expression of Bax and PUMA was significantly reduced, while the expression of Bcl-2 was significantly increased after co-transfection (Figure 6D). As expected, the migration and invasion capacities of T24 and SW780 cells was significantly promoted after miR-1290 mimic (Figure 6E and F).
miR-1290 Targets and Negatively Regulates CDC73
Subsequently, we downloaded the BC gene expression microarray GSE86411 from the GEO database, which contains 89 BC tissues and 43 normal bladder tissues, and we found a total of 621 mRNAs (Figure 7A and B) that were differentially expressed. Six genes (Figure 7C) were filtered by intersecting the prediction results of the target mRNAs of miR-1290 using TargetScan as well as the miRDB website and the poorly expressed mRNAs in the GSE86411 microarray. We put our attention on CDC73. We further tested the expression of CDC73 in cancerous and adjacent tissues of 57 BC patients. The expression of CDC73 in cancerous tissues was significantly lower than that in adjacent tissues (Figure 7D). Moreover, the expression of CDC73 was positively correlated with circ_0000629 and negatively correlated with miR-1290 (Figure 7E and F). We further examined the expression CDC73 in bladder uroepithelial cells and BC cells and observed that CDC73 expression was significantly reduced in the BC cell lines (Figure 7G). The expression of CDC73 increased at both mRNA and protein levels after overexpression of circ_000629, but decreased notably after further overexpression of miR-1290 in T24 and SW780 cells (Figure 7H and I). To further clarify the mechanism of regulation of CDC73 by miR-1290, we used dual-luciferase experiments. The luciferase activity in 293T cells transfected with miR-1290 mimic and CDC73-wt was significantly lower than that in cells transfected with mimic NC and CDC73-mt (Figure 7J).
Depletion of CDC73 Attenuates the Repressive Effect of oe-circ_0000629 on BC Cells
To investigate the role of CDC73 in BC, we further transfected two short hairpin RNAs (shRNAs) targeting CDC73 into T24 and SW780 cells overexpressing circ_0000629, and verified the knockdown efficiency of shRNAs on CDC73 by RT-qPCR and Western blot (Figure 8A and B). The growth of T24 and SW780 cells was significantly enhanced in the presence of oe-circ_0000629 + shCDC73 (Figure 8C). The significant increase in the number of apoptotic cells due to oe-circ_0000629 was partially reversed upon knockdown of CDC73 expression (Figure 8D). The protein expression of Bax and PUMA was significantly decreased, while the expression of Bcl-2 was significantly increased (Figure 8E). Moreover, we further found that the migration and invasion of T24 and SW780 cells were significantly promoted following the oe-circ_0000629 + shCDC73 treatment (Figure 8F and G).
Overexpression of Circ_0000629 Inhibits the Growth of BC Cells in vivo
To further evaluate the effect of circ_0000629 on BC cell growth in vivo, we inoculated T24 and SW780 cells stably overexpressing circ_0000629 or Vector NC into the axilla of nude mice by subcutaneous injection. Overexpression of circ_0000629 significantly inhibited the growth rate of tumors formed by T24 and SW780 cells in nude mice (Figure 9A and B). Subsequently, we used immunohistochemistry to detect the positive staining intensity of KI67 in tumors, and we noted that the staining intensity of KI67 in tumor tissues was significantly reduced after overexpression of circ_0000629 (Figure 9C).
Discussion
BC contributes to approximately 80,500 new diagnoses and 32,900 deaths in 2015 in China.13 In the meantime, circRNAs have been indicated to be dysregulated in multiple cancer tissues relative to matched normal tissues, especially in BC, signifying their role as diagnostic and prognostic biomarkers of BC.14,15 In the present work, a new circRNA, circ_0000629, was found remarkably downregulated in BC tissues and cells. We further revealed that circ_0000629 served as a tumor suppressor and had the potency to suppress the proliferation and aggressiveness of BC cells in vitro and in vivo. These findings exhibited that circ_0000629 may be involved in the development of BC and be an attractive therapeutic option.
Firstly, RT-qPCR and microarray analysis found circ_0000629 was poorly expressed in BC tissues and cell lines. Moreover, circ_0000629 upregulation led to repressed BC cell proliferation and accelerated apoptosis, as evidenced by elevated Caspase-3/7 activity, PUMA, and Bax expression, while reduced Bcl-2 expression. In addition, circ_0000629 was revealed to halt the processes of migration, invasiveness as well as EMT. EMT is a multistep event, during which epithelial cells acquire mesenchymal characteristics to support cancer cell motility and invasiveness.16 The epithelial cell surface markers include the most well-known factor E-cadherin, occluding and claudins, while the mesenchymal cell surface markers are comprised of N-cadherin, fibronectin as well as Vimentin.17 We observed by RT-qPCR that circ_0000629 restored E-cadherin expression, whereas decreased expression patterns of Vimentin, Snail and N-cadherin. However, the underlying mechanism is undetermined.
CircRNAs have also been reported to govern gene mediation by binding and preventing miRNAs from modulating their mRNA targets.18 For example, bladder cancer-related circRNA-3 could bind to miR-182-5p in a direct manner, and play as a miRNA sponge to enhance the miR-182-5p-targeted p27 expression in BC.19 While circFAM114A2 sequestrated miR-762 in modulating ΔNP63 expression, thereby suppressing BC progression.20 Nevertheless, information regarding miRNA sponging function of circ_0000629 is still limited. Herein, we confirmed that circ_0000629 was localized in cytoplasm with FISH assay and found that circ_0000629 contained binding sites with miR-1290 (an oncogene). The interaction between circ_0000629 and miR-1290 in BC was further identified by dual-luciferase experiment. Therefore, we could conclude that circ_0000629 may exert its anti-tumor properties in BC by binding to miR-1290.
In line with our findings, a comprehensive comparative analysis of GEO datasets conducted by Wei et al disclosed that miR-1290 was upregulated in pancreatic cancer patients and considerably decreased after tumor resection, suggesting its promising biomarker role for the diagnosis of pancreatic cancer.21 Kozinn et al has proposed that miR-1290 displayed increased expression in gemcitabine-resistant BC cells.22 Also, Khalighfard et al established that miR-1290 upregulation increased glioblastoma cell proliferation, migration, in addition to invasion.23 In the current study, miR-1290 was found to be notably upregulated in BC cells and tissue specimens, which was conversely correlated with circ_0000629 expression. Functional researches suggested that miR-1290 could facilitate BC development by enhancing cell aggressiveness of BC cells even in the presence of oe-circ_0000629. Lately, miR-1290 has been revealed to promote cancer aggressiveness in pancreatic ductal adenocarcinoma by directly targeting IKK1.24 Bioinformatics prediction and microarray analyses in our study revealed that miR-1290 could bind to CDC73 3ʹUTR. CDC73 functions as a tumor suppressor, and encodes parafibromin, a 531-amino acid protein mainly expressed in the nucleus.25 CDC73 was documented to suppress colony formation and cellular proliferation and induce cell cycle arrest in the G1 phase, suggesting its critical role in cell growth and proliferation.26 Furthermore, restoration of CDC73 level could be a prolific strategy to treat oral squamous cell carcinoma.27 Intriguingly, circ_0000140 has been identified as a tumor suppressor circRNA in oral squamous cell carcinoma by upregulating CDC73 via sequestration of miR-182-5p.28 Furthermore, silencing of CDC73 could negate the suppressive effects of oe-circ_0000629 on cell migration, invasion, and proliferation of BC in our study. Basis on these findings, we exhibited that circ_0000629 serves as a miRNA sponge to restrain the BC development through the circ_0000629/miR-1290/CDC73 axis.
Conclusion
In conclusion, the circ_0000629 is poorly expressed in BC tissues and cell lines. Overexpression of circ_0000629 represses migration, invasion and growth in BC cells through the circ_0000629/miR-1290/CDC73 axis. Nevertheless, whether our proposition is correct or whether there are other possible mechanisms has not been elucidated in this study, which awaits to be further explored.
Data Sharing Statement
All the data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
This study was approved by the Research and Ethical Committee of Hunan Provincial People’s Hospital and conducted in accordance with the Declaration of Helsinki. Participants signed informed consents prior to sample collection. Experimental procedures were approved by the Institutional Committee on the Care and Use of Animals of Hunan Provincial People’s Hospital. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no potential conflicts of interest.
References
1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi:10.1016/j.eururo.2016.06.010
2. Cumberbatch MGK, Noon AP. Epidemiology, aetiology and screening of bladder cancer. Transl Androl Urol. 2019;8(1):5–11. doi:10.21037/tau.2018.09.11
3. DeGeorge KC, Holt HR, Hodges SC. Bladder cancer: diagnosis and treatment. Am Fam Physician. 2017;96(8):507–514.
4. Anastasiadis A, de Reijke TM. Best practice in the treatment of nonmuscle invasive bladder cancer. Ther Adv Urol. 2012;4(1):13–32. doi:10.1177/1756287211431976
5. Li Y, Li G, Guo X, Yao H, Wang G, Li C. Non-coding RNA in bladder cancer. Cancer Lett. 2020;485:38–44. doi:10.1016/j.canlet.2020.04.023
6. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
7. Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell. 2014;159(1):13–14. doi:10.1016/j.cell.2014.09.005
8. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881 e813. doi:10.1016/j.cell.2018.12.021
9. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
10. Li Y, Qiao L, Zang Y, Ni W, Xu Z. Circular RNA FOXO3 suppresses bladder cancer progression and metastasis by regulating MiR-9-5p/TGFBR2. Cancer Manag Res. 2020;12:5049–5056. doi:10.2147/CMAR.S253412
11. Wu Y, Wei J, Zhang W, Xie M, Wang X, Xu J. Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. Onco Targets Ther. 2020;13:7809–7818. doi:10.2147/OTT.S263934
12. Li B, Wang F, Li X, Sun S, Shen Y, Yang H. Hsa_circ_0008309 may be a potential biomarker for oral squamous cell carcinoma. Dis Markers. 2018;2018:7496890. doi:10.1155/2018/7496890
13. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
14. Li C, Fu X, He H, Chen C, Wang Y, He L. The biogenesis, functions, and roles of circRNAs in bladder cancer. Cancer Manag Res. 2020;12:3673–3689. doi:10.2147/CMAR.S245233
15. Xie F, Zhao N, Zhang H, Xie D. Circular RNA CircHIPK3 promotes gemcitabine sensitivity in bladder cancer. J Cancer. 2020;11(7):1907–1912. doi:10.7150/jca.39722
16. Monteiro-Reis S, Lobo J, Henrique R, Jeronimo C. Epigenetic mechanisms influencing epithelial to mesenchymal transition in bladder cancer. Int J Mol Sci. 2019;20(2):297. doi:10.3390/ijms20020297
17. Ashrafizadeh M, Hushmandi K, Hashemi M, et al. Role of microRNA/epithelial-to-mesenchymal transition axis in the metastasis of bladder cancer. Biomolecules. 2020;10(8):1159. doi:10.3390/biom10081159
18. Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–2246. doi:10.1093/bioinformatics/btu257
19. Xie F, Li Y, Wang M, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17(1):144. doi:10.1186/s12943-018-0892-z
20. Liu T, Lu Q, Liu J, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating NP63 by sponging miR-762. Cell Death Dis. 2020;11(1):47. doi:10.1038/s41419-020-2226-5
21. Wei J, Yang L, Wu YN, Xu J. Serum miR-1290 and miR-1246 as potential diagnostic biomarkers of human pancreatic cancer. J Cancer. 2020;11(6):1325–1333. doi:10.7150/jca.38048
22. Kozinn SI, Harty NJ, Delong JM, et al. MicroRNA profile to predict gemcitabine resistance in bladder carcinoma cell lines. Genes Cancer. 2013;4(1–2):61–69. doi:10.1177/1947601913484495
23. Khalighfard S, Kalhori MR, Haddad P, Khori V, Alizadeh AM. Enhancement of resistance to chemo-radiation by hsa-miR-1290 expression in glioblastoma cells. Eur J Pharmacol. 2020;880:173144. doi:10.1016/j.ejphar.2020.173144
24. Ta N, Huang X, Zheng K, et al. miRNA-1290 promotes aggressiveness in pancreatic ductal adenocarcinoma by targeting IKK1. Cell Physiol Biochem. 2018;51(2):711–728. doi:10.1159/000495328
25. Korpi-Hyovalti E, Cranston T, Ryhanen E, et al. CDC73 intragenic deletion in familial primary hyperparathyroidism associated with parathyroid carcinoma. J Clin Endocrinol Metab. 2014;99(9):3044–3048. doi:10.1210/jc.2014-1481
26. Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics. J Biol Chem. 2013;288(1):608–618. doi:10.1074/jbc.M112.425736
27. Rather MI, Swamy S, Gopinath KS, Kumar A. Transcriptional repression of tumor suppressor CDC73, encoding an RNA polymerase II interactor, by Wilms tumor 1 protein (WT1) promotes cell proliferation: implication for cancer therapeutics. J Biol Chem. 2014;289(2):968–976. doi:10.1074/jbc.M113.483255
28. Guo J, Su Y, Zhang M. Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p. Cancer Cell Int. 2020;20:407. doi:10.1186/s12935-020-01501-7
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.