Back to Journals » Cancer Management and Research » Volume 12
Circular RNA MAT2B Induces Colorectal Cancer Proliferation via Sponging miR-610, Resulting in an Increased E2F1 Expression
Received 25 February 2020
Accepted for publication 19 June 2020
Published 10 August 2020 Volume 2020:12 Pages 7107—7116
DOI https://doi.org/10.2147/CMAR.S251180
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Jian Pei Zhao,1 Li Li Chen2
1Department of Anus & Intestine Surgery, HwaMei Hospital, University of Chinese Academy of Sciences, Ningbo 315010, Zhejiang, People’s Republic of China; 2Department of Hematology and Oncology, The First People’s Hospital of Taizhou, Taizhou 318020, Zhejiang, People’s Republic of China
Correspondence: Li Li Chen
Department of Hematology and Oncology, The First People’s Hospital of Taizhou, No. 218, Hengjie Road, Huangyan District, Taizhou, Zhejiang Province, People’s Republic of China
Tel +86 15105868468
Email [email protected]
Purpose: Recently, studies have demonstrated that a novel circular RNA (circRNA), circMAT2B, can promote cell proliferation and can thus contribute to the growth and development of hepatocellular carcinoma. However, the precise mechanisms underlying in circMAT2B-induced colorectal cancer (CRC) cell proliferation are not yet fully understood.
Materials and Methods: Quantitative reverse transcription polymerase chain reaction was conducted to evaluate circMAT2B expression in 70 CRC tissues and 70 matched adjacent normal tissues, CRC cell lines and human colonic epithelial cell line (NCM460). The direct interaction between miR-610 and circMAT2B or E2F1 was verified using luciferase reporter assay and biotinylated RNA Pull-down assay. Cell Counting Kit-8, colony formation assay, flow cytometry were utilized to examine the effect of circMAT2B, miR-610 and E2F1 on cell proliferation. Western blot was conducted to evaluate E2F1 expression.
Results: In our study, circMAT2B was found to be upregulated in CRC tissues and cell lines. Furthermore, the silencing of circMAT2B significantly inhibited proliferation. Hence, in order to investigate the mechanism underlying the oncogenic properties of circMAT2B in CRC, a bioinformatics analysis (circular RNA Interactome, https://circinteractome.nia.nih.gov/) was performed to screen the putative interacting microRNAs of circMAT2B. miR-610 was identified to be one of the potential targeted miRNAs of circMAT2B. Luciferase reporter and RNA pulldown assay confirmed a direct interaction between circMAT2B and miR-610. Moreover, circMAT2B expression was negatively correlated with the expression of miR-610 in CRC tissues (r=− 0.5131, p< 0.0001). Furthermore, we demonstrated circMAT2B upregulated expression levels of the miR-610 target gene E2F1, which is involved in cell proliferation, is overexpression in a broad range of human cancer including CRC. Further studies suggested that E2F1 upregulation could significantly reverse the si-circMAT2B-mediated inhibition of proliferation.
Conclusion: circMAT2B is upregulated in CRC tissues and cell lines. Moreover, circMAT2B promoted CRC proliferation by regulating the miR-610/E2F1 axis, which may serve as a potential therapeutic target for CRC treatment.
Keywords: circMAT2B, miR-610, E2F1, colorectal cancer, proliferation
Introduction
Among malignant tumors, the incidence of colorectal cancer (CRC) is the second in males and third in females.1 The preferred treatment for patients with colorectal cancer is surgical resection followed by adjuvant chemotherapy or radiotherapy. However, 30%–50% of patients undergoing curative resection have local and systemic recurrence, which is the leading cause of death in many colorectal cancer patients.2 Therefore, we need to find new therapeutic strategies, especially those that target disease-related molecular.
With the increasing number of genome-wide expression analysis research, a variety of circular RNAs (circRNAs) have gradually been identified, and these circRNAs have altered expression patterns in cancers including CRC.3 circRNAs is a new type of non-coding RNAs, which do not have 5′ caps and 3′ poly(A) tails, and form a closed circular structure through covalent bonds. Compared with linear RNA, circRNAs are more stable, abundant, conserved due to its special circular structure.4,5 Recently, studies have demonstrated that a novel circRNA, circMAT2B, can contribute to cell proliferation, thereby promoting the growth of hepatocellular carcinoma (HCC).6 However, the precise mechanisms of circMAT2B-induced CRC cell proliferation are unknown.
As is known, circRNAs can alter its own expression by sponging miRNAs or binding to proteins, resulting in abnormal expression of gene products, which may promote the occurrence of cancer.7,8 miR-610, located at 11p14.1, was a newly identified functional miRNA in tumors. It has been reported that the miR-610 inhibited the expression of CCND2 and AKT3, thereby repressing the proliferation of human glioblastoma cells.9 Studies have shown that the expression of miR-610 is significantly down-regulated in glioma cells, and miR-610 can block cell proliferation and metastasis by directly targeting MDM2.10 Reportedly, E2F transcription factor 1 (E2F1) is a member of the E2F family and plays an important role in controlling the cell cycle.11 In addition, E2F1 preferentially binds to the retinoblastoma protein pRB in a cell cycle-dependent manner and mediates both cell proliferation and p53-dependent/independent apoptosis. More and more studies show that this regulatory pathway is abnormal in many human malignancies.12
This study showed that the expression of circMAT2B was significantly higher in CRC tissues than that in adjacent tissues. Furthermore, a negative correlation was found between the expression of circMAT2B and miR-610. CircMAT2B downregulates the level of miR-610 target gene E2F1, which mediate cell proliferation, through sponging miR-610, thus promoting CRC progression. Therefore, circMAT2B is considered as a promising target for CRC treatment.
Materials and Methods
Human Specimens
This study was approved by the Ethics Committee of the First People’s Hospital of Taizhou and written informed consents were obtained from all patients. This was conducted in accordance with the Declaration of Helsinki. The paired tissue samples from the surgical specimens of the First People’s Hospital of Taizhou (Taizhou, China). In the study, the samples were stored using liquid nitrogen. Seventy CRC tissues and 70 adjacent normal colorectal tissues were collected.
Cell Lines
Seven human CRC cell lines including NCM460, HT29, HCT116, HCT8, SW480, DLD1, Caco2 were provided by the ATCC (American Type Culture Collection, Manassas, VA, USA). The cells were cultured in 1640 medium with 10% FBS (Gibco, Grand Island, NY, USA) at 37°C in an incubator containing 5% CO2.
Oligonucleotide Transfection
The scrambled control, miR-610 mimic, miR-610 inhibitor, siRNA targeting circMAT2B and siRNA negative control were all produced by GenePharma Co. (Shanghai, China). The cDNA of E2F1 was subcloned into pcDNA3.1 vector by RiboBio (Guangzhou, China). HCT116 and HT29 cells (1 × 105 cells/well) were seeded in 6-well plates and then transfected with Lipofectamine 3000 from Invitrogen.
Quantitative Reverse Transcription Polymerase Chain Reaction (qPCR)
Using TRIzol reagent from Thermo Fisher Scientific, total RNA was extracted from CRC tissues and cells, then using the reverse transcription reagent kit from Vazyme Biotech (Nanjing, China), the cDNA was generated. In a 20 µL reaction system, the qPCR was performed according to the instructions. Ten-microliter 2X qPCR SYBR-Green master mix (Vazyme Biotech), 0.4 µL forward primer (10 µM), 0.4 µL reverse primer (10 µM), 1 µL cDNA and 0.4 µL 50X ROX reference dye 1 were included in the 20 µL reaction system. All specimens were tested in triplicate. All primers used in this study are listed in Table 1. The results were corrected by the expression of β-actin and the 2−∆∆Cq method was used for the analysis of results.13
 |
Table 1 Specific Primer for qPCR |
Cell Counting Kit-8 (CCK-8) and Colony Formation Assay
In a 96-well plate, 2 × 103 cells were seeded in each well. After cells and 10 μL CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) were incubated at 37°C for 1 hr, the absorbance of the mixed solution at 450 nm was measured. In a 6-well plate, HCT116 and HT29 cells (500 per well) were seeded and cultured for 1–2 weeks. The colonies were stained with crystal violet solution and counted with a microscope. All tested were repeated three times.
Flow Cytometry
At 48 hrs after transfection, cells were collected and stained with Annexin V-Alexa Fluor 647 and PI (Fcmacs, Nanjing, China) for 15 mins. The processed samples were tested by Beckman Cytoflex flow cytometer. Using CytExpert software, the percentage of apoptosis cells was analyzed.
Luciferase Reporter Assay
CircMAT2B sequences containing wild-type or mutant miR-610 binding sites were synthesized. The sequences were then inserted into psi-CHECK2 vector (Promega, Madison, WI, USA). Then, the wild-type or mutated vector and miR-610 mimics or mimics control were co-transfected into HCT116 and HT29 cells using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA), respectively. 48h after transfection, the luciferase activity of the collected cells was detected by the Dual-Luciferase Reporter Assay System from Promega.
Biotinylated RNA Pull-Down Assay
The pull-down assay with biotinylated RNA was operated according to the previous process.14,15 In short, in order to pull down miRNAs for circMAT2B, probe-coated beads were generated by co-cultivation of the biotinylated-circMAT2B probe and C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA). Then, the probe-coated beads and sonicated HCT116 or HT29 cells were co-incubated at 4°C overnight, followed by elution and qPCR.
Western Blot
Samples were lysed in radioimmunoprecipitation buffer (Solarbio) to extract total protein. The protein concentration was detected by a bicinchoninic acid protein assay kit. The extracted total proteins were separated using SDS-PAGE and then transferred onto polyvinylidene fluoride membranes (Merck KGaA, Darmstadt, Germany). The membranes were incubated with primary antibodies, and then with a secondary antibody. Protein detection was performed using an enhanced chemiluminescence substrate kit from Thermo Scientific and ImageJ software.
Statistical Analysis
SPSS 22.0 software was used for statistical analysis, and GraphPad Prism 6.0 was used for producing figures. All experimental data are expressed in the form of mean ± S.D, and calculated by Student’s t-test as appropriate. Probability (P) values <0.05 were considered statistically significant.
Results
Elevated circMAT2B Expression Levels in CRC Tissue and Cell Lines
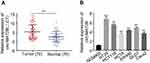
At the beginning of the study, we tested the expression level of circMAT2B in CRC tissues and cell lines, respectively. Using qPCR, we found that circMAT2B is significantly higher in the 70 cases of CRC (the clinical characteristics of the patients are summarized in Table 2) than in matched normal tissues (Figure 1A). Similar results were observed in CRC cell lines compared with NCM460 human colonic epithelial cells. Statistical analyses showed that CircMAT2B was markedly overexpressed in the HT29, HCT116, HCT8, SW480, DLD1, Caco2 cells, particularly in the HT29 and HCT116 cells (Figure 1B).
 |
Table 2 The Characteristics of the Patients with Colorectal Cancer (CRC) |
circMAT2B Knockdown Inhibits CRC Cells Proliferation
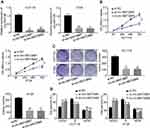
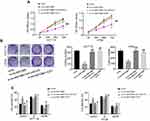
To explore the function of circMAT2B in the CRC process, we assessed the expression of circMAT2B in different CRC cell lines. Hence, we planned to knockdown the expression of circMAT2B in HCT116 and HT29 cell lines. Two siRNA targeting circMAT2B were designed and qPCR showed that these two siRNAs significantly decreased circMAT2B level (Figure 2A). Due to the highest efficiency of interference, si-circMAT2B#1 and si-circMAT2B#2 were used for the subsequent experiments. Silencing circMAT2B significantly inhibited the proliferation of CRC cell lines (Figure 2B-D).
circMAT2B Directly Binds to miR-610 in CRC
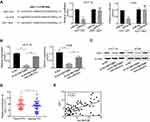
More and more studies have confirmed that circRNAs can act as ceRNAs to sponge miRNAs.16–18 Therefore, to further explore the potential carcinogenic mechanism of circMAT2B in CRC, a bioinformatics analysis (circular RNA Interactome, https://circinteractome.nia.nih.gov/) was performed to screen the putative interacting microRNAs of circMAT2B. miR-610 was identified to be one of the potential targeted miRNAs of circMAT2B (Figure 3A). To validate the direct interaction between miR-610 and circMAT2B, a dual-luciferase reporter assay was conducted by co-transfecting the recombinant plasmid containing the putative or mutant binding sites (Figure 3A) with miR-610 mimics or its negative control (miR-NC). The results showed that only the luciferase activity of wild-type circMAT2B could be attenuated by the miR-610 mimics (Figure 3A). Furthermore, biotin-coupled circMAT2B pulldown assay confirmed that miR-610 was found in the circMAT2B pulled down pellet compared with the biotin-coupled oligo group (Figure 3B), which indicates that circMAT2B could directly sponge miR-610. In addition, the expression of miR-610 was eminently upregulated when circMAT2B was silenced (Figure 3C). qPCR also confirmed that the expression of miR-610 was significantly lower in CRC tissues than that in adjacent normal tissues (Figure 3D). In addition, circMAT2B expression in CRC tissues was negatively correlated with the expression of miR-610 (r=−0.5131, p<0.0001) (Figure 3E). Taken together, these results demonstrate that circMAT2B directly binds to miR-610 in CRC.
circMAT2B Increases the Expression of Oncogene E2F1 by Sponging miR-610 in CRC
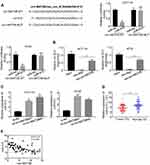
There is increasing evidence that miR-610 acts on multiple oncogenes simultaneously. Therefore, we explored that circMAT2B promotes tumor proliferation by regulating the expression of miR-610 targeting oncogenes. Expected targets were determined using the bioinformatics tool miRanda. The analysis revealed that E2F1 3ʹ-UTR has miR-610 binding sites (Figure 4A). To confirm that miR-610 regulated E2F1, a luciferase-based reporter gene plasmid with WT or mutated 3ʹ-UTR sequences for E2F1 was constructed. The luciferase activity of E2F1 was significantly reduced when cells were co-transfection with miR-610 mimics and reporter plasmids. However, there was no evident effect on the luciferase activity in cells transfected with miR-610 mimics and mutated vectors (Figure 4A). In HCT116 and HT29 cells with circMAT2B knockdown, transcription of E2F1 was significantly decreased, and restored by using miR-610 inhibitor (Figure 4B). The above data indicated that miR-610 reduces the mRNA expression of E2F1 by targeting its 3ʹ-UTR. Consistent with transcription data, Western blot analysis showed that E2F1 was regulated by circMAT2B and miR-610 in CRC cell lines (Figure 4C). Furthermore, qPCR also showed that E2F1 expression was significantly higher in CRC tissues than that in adjacent normal tissues (Figure 4D). Moreover, the expression of E2F1 was positively correlated with circMAT2B expression in CRC tissues (r=−0.5131, p<0.0001) (Figure 4E). These results confirmed that E2F1 expression is regulated by miR-610 bound to its 3ʹUTR, and miR-610 is sponged by circMTA2B.
Silencing of miR-610 or Overexpression of E2F1 Remarkably Rescues the Decreased Proliferative Ability of CRC Cells Caused by circMAT2B Silencing
We further examined whether circMAT2B regulates the expression of oncogene E2F1 by sponging miR-610. First, the HCT116 and HT29 cells were transfected with si-NC, si-circMAT2B#1, si-circMAT2B#1+inh-miR-610 and si-circMAT2B#1+E2F1. Next, CCK-8 assays, colony formation assays, and flow cytometry were used for the analysis of CRC proliferation. The data showed that the silencing of miR-610 or overexpression of E2F1 reversed the inhibiting effect of circMAT2B silencing on cell proliferation (Figure 5A-C) in CRC.
Discussion
For the past 30 years, circRNAs have been considered as noncoding RNAs (ncRNAs) and ignored as transcriptional noise of eukaryotes.19,20 However, due to the rapid development of bioinformatics, the widespread expression of circRNAs has been found in a variety of cells and species.21 Moreover, more and more scientists recognize that circRNAs may play a vital role in the development of cancer and affect the hallmarks of cancer.16–18,22–25 However, the specific function of circRNAs in the development of CRC is still unclear. In this study, qPCR detected the expression of circMAT2B in 70 pairs of CRC and adjacent tissues, and the results showed that circMAT2B was significantly higher in CRC tissues. Moreover, negative correlations were also found between the expression of circMAT2B and miR-610. CircMAT2B downregulates the level of miR-610 target gene E2F1, which mediate cell proliferation, through sponging miR-610, thus promoting CRC progression. These results indicated that circMAT2B might be related to the progression of CRC.
Research has shown that ubiquitin-like modifier-activating enzyme 2 (UBA2) expression may enhance CRC cell proliferation. Furthermore, the expression of UBA2 may be associated with p53/MDM2/p21 and PI3K/AKT signaling pathways.26 Another study confirmed that miR-203a inhibits CRC proliferation by targeting RING-finger protein 6 (RNF6), offering novel insights into the regulatory network of miR-203a-modulated cell cycle and proliferation, and suggest that miR-203a was a potential therapeutic target in CRC treatment.27 To explore the diagnostic values of circMAT2B in CRC, qPCR was used to measure circMAT2B expression in 70 pairs of CRC and matched adjacent tissues. The results confirmed that compared with matched adjacent tissues, the mRNA level of circMAT2B was significantly increased in CRC tissues. Moreover, the functional studies in CRC cell lines showed that circMAT2B promotes CRC cell proliferation, suggesting that circMAT2B might be related to the development of CRC and could become a promising therapeutic target for CRC. Consistent with this, a previous study has shown a significant increase in circMAT2B expression in HCC tissues and cell lines, and increased circMAT2B indicated poor prognosis, suggesting its tumor-promoting effect.6 The underlying mechanisms of circRNA function in cancer development and progression have not been clarified.28 The most widely reported function of circRNAs is as miRNA sponges in the circRNA–miRNA–mRNA axes.29,30 In order to study the mechanism of the oncogenic properties of circMAT2B in CRC, the target miRNAs of circMAT2B was predicted by circular RNA Interactome software. The results suggested that circMAT2B contains a putative binding site for miR-610 based on the bioinformatics analysis. Furthermore, we confirmed that circMAT2B could sequester and inhibit miR-610 activity by sponging endogenous miR-610, which targets the oncogene E2F1, which is related to cell proliferation, and is overexpression in a variety of human cancer including CRC.31–33 Further studies showed that the down-regulation of miR-610 and the upregulation of E2F1 could effectively reverse the si-circMAT2B-mediated inhibition of proliferation in HCT116 and HT29 cells. Overall, our research confirmed that circMAT2B promoted CRC proliferation by sponging miR-610, which targets the oncogene E2F1.
In conclusion, oncogenic circMAT2B was identified, and the role and mechanism in CRC proliferation was investigated. All the above data showed that circMAT2B expression was significantly upregulated in CRC tissues and cell lines. Moreover, circMAT2B could be used as an effective non-invasive biomarker for CRC detection. CircMAT2B accelerated CRC proliferation by regulating the miR-610/E2F1 axis, which may serve as a promising therapeutic target for CRC treatment.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Zhang L, Zheng C, Sun Z, Wang H, Wang F. Long non-coding RNA urothelial cancer associated 1 can regulate the migration and invasion of colorectal cancer cells (SW480) via myocardin-related transcription factor-A. Oncol Lett. 2019;18(4):4185–4193. doi:10.3892/ol.2019.10737.
2. Imai J, Koganezawa Y, Tuzuki H, Ishikawa I, Sakai T. An optical and non-invasive method to detect the accumulation of ubiquitin chains. Cell Biol Int. 2019;43(12):1393–1406. doi:10.1002/cbin.11186.
3. Galamb O, Bartak BK, Kalmar A, et al. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol. 2019;25(34):5026–5048. doi:10.3748/wjg.v25.i34.5026.
4. Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332. doi:10.3389/fonc.2018.00332.
5. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112.
6. Li Q, Pan X, Zhu D, Deng Z, Jiang R. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi:10.1002/hep.30671.
7. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi:10.1158/0008-5472.can-13-1568.
8. Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. doi:10.1038/onc.2017.89.
9. Mo X, Cao Q, Liang H, Liu J, Li H, Liu F. MicroRNA-610 suppresses the proliferation of human glioblastoma cells by repressing CCND2 and AKT3. Mol Med Rep. 2016;13(3):1961–1966. doi:10.3892/mmr.2016.4760.
10. Yan Y, Peng Y, Ou Y, Jiang Y. MicroRNA-610 is downregulated in glioma cells, and inhibits proliferation and motility by directly targeting MDM2. Mol Med Rep. 2016;14(3):2657–2664. doi:10.3892/mmr.2016.5559.
11. Castillo DS, Campalans A, Belluscio LM, et al. E2F1 and E2F2 induction in response to DNA damage preserves genomic stability in neuronal cells. Cell Cycle. 2015;14(8):1300–1314. doi:10.4161/15384101.2014.985031.
12. Dubrez L. Regulation of E2F1 transcription factor by ubiquitin conjugation. Int J Mol Sci. 2017;18(10):2188. doi:10.3390/ijms18102188.
13. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262.
14. Li Y, Zheng F, Xiao X, et al. Circ HIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi:10.15252/embr.201643581.
15. Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi:10.1093/eurheartj/ehv713
16. Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78(17):4812–4825. doi:10.1158/0008-5472.can-18-0532.
17. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi:10.1002/hep.29270.
18. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027.
19. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559.
20. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping.. Proc Natl Acad Sci U S A. 1996;93(13):6536–6541. doi:10.1073/pnas.93.13.6536.
21. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928.
22. Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–4015. doi:10.7150/thno.24106.
23. Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–3928. doi:10.1158/1078-0432.ccr-16-2541.
24. Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi:10.1016/j.canlet.2016.12.006.
25. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361.
26. He P, Sun X, Cheng HJ, et al. UBA2 promotes proliferation of colorectal cancer. Mol Med Rep. 2018;18(6):5552–5562. doi:10.3892/mmr.2018.9613.
27. Miao J, Hou N, Yang W, et al. miR-203a suppresses cell proliferation by targeting RING-finger protein 6 in colorectal cancer. Anticancer Drugs. 2020;31:583–591. doi:10.1097/cad.0000000000000874.
28. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034.
29. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. doi:10.1007/s00432-016-2256-7.
30. Tang W, Ji M, He G, et al. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco Targets Ther. 2017;10:2045–2056. doi:10.2147/ott.s131597.
31. Qian W, Zhang Z, Peng W, et al. CDCA3 mediates p21-dependent proliferation by regulating E2F1 expression in colorectal cancer. Int J Oncol. 2018;53(5):2021–2033. doi:10.3892/ijo.2018.4538.
32. Yin J, Shen X, Li M, Ni F, Xu L, Lu H. miR-329 regulates the sensitivity of 5-FU in chemotherapy of colorectal cancer by targeting E2F1. Oncol Lett. 2018;16(3):3587–3592. doi:10.3892/ol.2018.9121.
33. Li SZ, Zeng F, Li J, et al. Nemo-like kinase (NLK) primes colorectal cancer progression by releasing the E2F1 complex from HDAC1. Cancer Lett. 2018;431:43–53. doi:10.1016/j.canlet.2018.05.032.c
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.