Back to Journals » Cancer Management and Research » Volume 11
Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis
Authors Li R, Wu B, Xia J, Ye L, Yang X
Received 13 April 2019
Accepted for publication 28 June 2019
Published 23 July 2019 Volume 2019:11 Pages 6887—6893
DOI https://doi.org/10.2147/CMAR.S212180
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Rizeng Li,1 BinBin Wu,2 Jianfu Xia,1 Lechi Ye,3 Xiaoping Yang4
1Department of Colorectal Surgery, Wenzhou Central Hospital, Wenzhou 325000, People’s Republic of China; 2Department of General Surgery, The Second People’s Hospital of Pingyang County, Wenzhou 325000, People’s Republic of China; 3Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China; 4Department of Digestive Medicine, First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Purpose: Circular RNAs (circRNAs) are recently identified new noncoding RNAs and play an important role in tumorigenesis. Previous studies have indicated that hsa_circRNA_102958 is a potential diagnostic indicator in gastric cancer. However, its role in colorectal cancer (CRC) is poorly understood.
Methods: qRT-PCR and ISH were used to test gene expression in tissues. Survival rate was analzyed by Kaplan-Meier curve. Luciferase reporter assay was used to determine the interaction between circRNA and miRNA or between miRNA and mRNA. Western blotting was used to test protein expression. CCK8 and colony formation assay was used to analyze proliferation. Transwell assay was used for migration and invasion determination.
Results: In our research, we found that hsa_circRNA_102958 expression was significantly increased in CRC tissues, compared to adjacent normal controls. Increased hsa_circRNA_102958 levels in CRC patients indicated a poor prognosis. The effects of hsa_circRNA_102958 on CRC cell proliferation, migration and invasion were then determined by CCK8, colony formation and Transwell assays. We showed that hsa_circRNA_102958 silencing markedly suppressed CRC growth, migration and invasion. Furthermore, hsa_circRNA_102958 was identified as a sponge for miR-585. We demonstrated that hsa_circRNA_102958 promoted CDC25B expression through inhibiting miR-585 in CRC. Rescue assays illustrated that CDC25B overexpression reversed the suppressive effects of hsa_circRNA_102958 silencing on CRC.
Conclusion: Taken together, our findings revealed the novel oncogenic roles of hsa_circRNA_102958 in CRC through miR-585/CDC25B axis.
Keywords: hsa_circRNA_102958, miR-585, CDC25B, proliferation, invasion
Introduction
Based on the recent data, colorectal cancer (CRC) is the third most common cancer and causes a huge number of deaths.1 Although great advance has been obtained on strategies for CRC therapy, the mortality of CRC remains very high.2 The 5-year survival rate of CRC patients is very low.3 Thus, the molecular mechanism of CRC development requires to be understood and novel therapeutic targets are urgently waiting for determination.
Circular RNAs (circRNAs) are a novel kind of noncoding RNAs and characterized by a covalently closed loop and lacking 5ʹ caps and 3ʹ poly(A) tails.4 In the past years, increasing numbers of evidence have revealed the vital importance of circRNAs in human diseases, including cancers.5,6 Many biological processes, such as cell proliferation, metastasis, differentiation and death, could be regulated by circRNAs in cancer.7 For instance, circRNA cTFRC interacts with miR-107 to enhance the growth and metastasis of bladder cancer.8 CircPSMC3 competitively sponges miR-29605p to inhibit cell proliferation, migration and invasion in gastric cancer.9 CircRNA hsa_circ_0072309 represses growth and invasiveness of breast cancer through sponging miR-492.10 These researchers indicate the key regulatory roles of circRNAs in tumorigenesis. However, the mechanism of circRNA regulating CRC still remains poorly understood.
Hsa_circRNA_102958 is a novel circRNA and its function has not been well investigated. A recent report indicated that hsa_circRNA_102958 may be a diagnostic biomarker for gastric cancer.11 However, the function of hsa_circRNA_102958 in CRC is unclear. In this study, we found that hsa_circRNA_102958 expression was upregulated in CRC tissues and cell lines. Hsa_circRNA_102958 high expression in CRC patients indicated a poor prognosis. Silencing of hsa_circRNA_102958 significantly suppresses the proliferation, migration and invasion of CRC cells in vitro. Moreover, tumor growth of CRC was also inhibited by hsa_circRNA_102958 knockdown in vivo. Hsa_circRNA_102958 was demonstrated to interact with miR-585 and promote CDC25B expression mechanistically. In conclusion, we identified that hsa_circRNA_102958/miR-585/CDC25B axis is a novel and pivot signaling pathway involved in CRC progression.
Materials and methods
Patients and samples
All 58 patient samples were obtained from our hospital. Tissues were stored in liquid nitrogen after collection. No patients were treated by chemotherapy or radiotherapy. This study was approved by the Ethics Committee of Wenzhou Central Hospital. Informed consent was signed by all patients. Experiments involving human tissues were conducted in accordance with the Declaration of Helsinki.
Cell culture
CRC cell lines, including LoVo, SW480, SW620, HT29, HCT8 and HCT116 cells, and normal colon epithelial cell line FHC were obtained from Cell Bank of Type Culture Collection (Shanghai City, China). Cells were cultured using DMEM (Invitrogen) containing 10% FBS (Gibco,Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco).
Quantitative real-time PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Quantitative real-time PCR analysis was conducted as previously reported.5
Bioinformatics analysis
The potential target of hsa_circRNA_102958 was analyzed by using circinteractome (https://circinteractome.nia.nih.gov/). Potential targets of miR-585 were predicted by using TargetScan (http://www.targetscan.org/vert_71/).
Luciferase reporter assay
The wild type (WT) or mutant sequence of hsa_circRNA_102958 or CDC25B was cloned into psiCHECK-2 vector (Promega, Madison, WI, USA). Then, reporters and miR-585 mimics or controls were transfected into CRC cells using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA); 48 hrs later, the relative luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase activity is internal control.
CCK8 assay
CRC cells were seeded into the 96-well plates at a density of 2000 cells per well. Then, cells were cultured for 24, 48 and 72 hrs. Later, the Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) was added and incubated for 4 hrs. Then, absorbance at 450 nm was then detected by using a Varioskan Flash Spectral Scanning Multimode Reader (Thermo Fisher Scientific, Waltham, MA, USA).
Transwell assay
Cell migration and invasion were evaluated by using Transwell chamber (Corning Inc., Corning, NY, USA) and Matrigel (Corning Inc.) as previously described.2
In vivo assay
Four-week-old male BALB/c nude mice were divided into 2 groups (n=5 for each group). Then, HCT116 cells (hsa_circRNA_102958 silencing or control) were subcutaneously inoculated into the right flank of each nude mouse. Every 1 week, the tumor volumes were measured by the formula: Volume = 1/2 (length × width2). This assay was approved by the Ethics Committee of Wenzhou Central Hospital. All animal operations were performed in accordance with the Animal Policy and Welfare Committee of Wenzhou Central Hospital.
Statistical analysis
GraphPad Prism 6.0 was used for Statistical analysis (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test or one-way ANOVA was used for calculating significant differences. Survival rate was measured by Kaplan–Meier method and log-rank test. The correlations were determined through Pearson’s correlation coefficients. All experiments were conducted at least three times. P< 0.05 was considered significantly different.
Results
Expression level of hsa_circRNA_102958 was upregulated in CRC
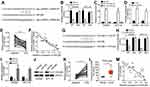
The expression of hsa_circRNA_102958 in CRC tissues was analyzed by quantitative real-time PCR. We found that hsa_circRNA_102958 level was increased in tumor tissues (n=58) compared to adjacent normal tissues (n=58) (Figure 1A). To further validate it, in situ hybridization was conducted and similar results were achieved (Figure 1B). Furthermore, we found that hsa_circRNA_102958 expression was positively correlated with clinical stage (Figure 1C) and lymph node metastasis (Figure 1D). Consistently, hsa_circRNA_102958 expression was also upregulated in CRC cell lines compared to FHC cells (Figure 1E). Then, the samples were divided into hsa_circRNA_102958 high expression and low expression groups based on the median value. Survival rate was calculated by Kaplan–Meier curve. Result showed that hsa_circRNA_102958 high expression group displayed a low survival rate (Figure 1F).
Loss of hsa_circRNA_102958 suppressed proliferation, migration and invasion of CRC cells
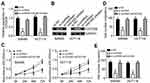
To investigate the potential roles of hsa_circRNA_102958, SW480 and HCT116 cells were selected for experiments because hsa_circRNA_102958 expression was the highest in these two cells among all detected cell lines (Figure 1E). Hsa_circRNA_102958 expression was decreased by using specific siRNAs (Figure 2A). Through CCK8 assay, we found that hsa_circRNA_102958 knockdown significantly suppressed the proliferation of SW480 and HCT116 cells (Figure 2B). Colony formation further validated it (Figure 2C). Transwell assay showed that hsa_circRNA_102958 knockdown inhibited the migration and invasion of SW480 and HCT116 cells (Figure 2D and E). To further confirm the effect of hsa_circRNA_102958 on CRC growth in vivo, we performed xenograft assay. We found that hsa_circRNA_102958 silencing significantly inhibited tumor volumes (Figure 2F). Thus, hsa_circRNA_102958 promotes proliferation, migration and invasion of CRC.
Hsa_circRNA_102958 was a sponge for miR-585 and miR-585 targeted CDC25B
Previous studies have shown that circRNA are good sponges for miRNAs.7,10 Thus, we analyzed the potential targets of hsa_circRNA_102958. Through bioinformatics analysis, we found that hsa_circRNA_102958 might be the sponge for miR-585 (miR-585-3p) because miR-585 ranked top among all candidates by bioinformatics analysis (Figure 3A). To validate it, we performed luciferase reporter assay. We found that the activity of WT hsa_circRNA_102958 reporter was suppressed by miR-585 mimics in SW480 and HCT116 cells (Figure 3B). Besides, RNA pulldown also indicated that biotin-labeled WT miR-585 could interact with hsa_circRNA_102958 (Figure 3C), suggesting their direct interaction. Notably, we observed that hsa_circRNA_102958 silencing led to increased expression of miR-585 in CRC cells (Figure 3D). Furthermore, we found that miR-585 expression was downregulated and negatively correlated with hsa_circRNA_102958 levels in CRC tissues (Figure 3E and F). Afterwards, we analyzed the potential targets of miR-585. According to bioinformatics analysis, we identified CDC25B because it ranked top and has been reported to participate in tumorigenesis (Figure 3G). Through luciferase reporter assay, we found that the activity of WT CDC25B-3ʹ UTR was significantly reduced by miR-585 mimics (Figure 3H). Moreover, miR-585 mimics inhibited the expression of CDC25B in CRC cells (Figure 3I and J) and miR-585 inhibitors promoted CDC25B expression (Figure 3I). We then analyzed the expression patterns of CDC25B. Quantitative real-time PCR indicated that CDC25B levels were upregulated in CRC tissues (Figure 3K), which was consistent with the TCGA data (Figure 3L). Interestingly, we observed that CDC25B expression was negatively correlated with miR-585 in CRC tissues (Figure 3M). Thus, above data demonstrates that miR-585 is sponged by hsa_circRNA_102958 and targets CDC25B in CRC.
Hsa_circRNA_102958 promoted CRC progression through CDC25B
To determine whether hsa_circRNA_102958 promotes CRC progression through CDC25B, we measured the effect of hsa_circRNA_102958 on CDC25B expression. We found that CDC25B expression was decreased by hsa_circRNA_102958 knockdown (Figure 4A and B), whereas inhibition of miR-585 reversed it (Figure 4A and B). Thus, hsa_circRNA_102958 promotes CDC25B expression by inhibiting miR-585. Then, we overexpressed CDC25B. As shown, CDC25B overexpression reversed the effects of hsa_circRNA_102958 knockdown on cell proliferation, migration and invasion (Figure 4C–E). Taken together, our results illustrate the key roles of hsa_circRNA_102958/miR-585/CDC25B axis in CRC.
Discussion
In the current study, we investigated the oncogenic roles of hsa_circRNA_102958 in CRC progression. We showed that hsa_circRNA_102958 expression was significantly upregulated in CRC tissues compared to adjacent normal tissues. Furthermore, hsa_circRNA_102958 level was positively correlated with clinical stage. And, increased expression of hsa_circRNA_102958 predicted poor prognosis. Loss-of-function assays indicated that hsa_circRNA_102958 silencing significantly suppressed the proliferation, migration and invasion of CRC cells. We also found that hsa_circRNA_102958 was a sponge for miR-585 and upregulated miR-585 expression.
Accumulating studies have demonstrated the essential functions of circRNAs in cancers, including CRC.2 Several evidences reveal that circRNAs participate in regulating various life processes of CRC cells, such as differentiation, proliferation and migration. For example, hsa_circ_0136666 increases the growth and metastasis of CRC by modulating miR-136/SH2B1 pathway.12 CircRNA_100290 contributes to CRC development via miR-516b/FZD4/Wnt/β-catenin signaling cascade.13 Overexpression of hsa_circ_0000069 enhances the growth and metastasis of CRC.14 However, the relationship between circRNA and CRC progression still remains poorly explored. And the roles of hsa_circRNA_102958 are largely unknown. In our study, we found that hsa_circRNA_102958 expression was significantly upregulated in CRC tissues. Through CCK8 and colony formation assays, we demonstrated that hsa_circRNA_102958 promotes CRC cell proliferation. By using Transwell assay, we revealed that hsa_circRNA_102958 silencing suppressed migration and invasion of CRC cells. Xenograft assay also demonstrated the oncogenic roles of hsa_circRNA_102958. Taken together, our work demonstrated that hsa_circRNA_102958 promoted CRC cell proliferation, migration and invasion in vitro and in vivo.
To further investigate the molecular mechanism, we used bioinformatics method to search the potential miRNA targets of hsa_circRNA_102958. We identified that hsa_circRNA_102958 possesses a potential binding site for miR-585. Through luciferase reporter assay and RNA pulldown assay, we demonstrated their direct interaction. We also observed that miR-585 expression was inhibited by hsa_circRNA_102958. miR-585 is a poorly researched miRNA. Two previous studies indicate that miR-585 is an anti-tumor gene in lung cancer and gastric cancer.15,16 Whether miR-585 is involved in CRC remains unknown. In our study, we found that miR-585 expression was significantly downregulated in CRC tissues. Our data also suggested that miR-585 is a tumor suppressor in CRC. Afterwards, we further investigated the target genes of miR-585. By bioinformatics analysis, we identified CDC25B as a potential target. Through luciferase reporter assay, we demonstrated that CDC25B was directly targeted by miR-585. Additionally, we showed that miR-585 mimics directly suppressed CDC25B expression in CRC. CDC25B is a cell-cycle regulator. Many literatures have demonstrated its oncogenic function. For example, CDC25B suppression by miR-152 leads to cell-cycle arrest of endometrial cancer cells.17 Cdc25B upregulation promotes cell cycle progression and contributes to melanoma.18 The role of CDC25B in CRC still remains unclear. In our study, we also demonstrated that CDC25B expression was upregulated by hsa_circRNA_102958 through inhibiting miR-585. Moreover, we showed that CDC25B expression was increased in CRC tissues. Additionally, restoration of CDC25B significantly rescued the proliferation, migration and invasion of hsa_circRNA_102958-silenced CRC cells. Thus, our findings demonstrated the oncogenic roles of CDC25B in CRC.
Notably, in the future study, whether circRNAs could be used for diagnosis or served as therapeutic targets should be researched in the future work. And our work also has some limitations. For example, we did not study the correlation between circRNA expression and clinical features.
In summary, we confirmed the high expression of hsa_circRNA_102958 in CRC tissues. We also demonstrated the oncogenic roles of hsa_circRNA_102958 by promoting proliferation, migration and invasion of CRC cells. Finally, we revealed a novel mechanism. We showed that hsa_circRNA_102958/miR-585/CDC25B axis participates in CRC progression.
Acknowledgment
This study was granted by Zhejiang Provincial Medical Technology Science Project (2015KYB245).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Li XN, Wang Z-J, Ye C-X, Zhao B-C, Huang X-X, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112:108611. doi:10.1016/j.biopha.2019.108611
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi:10.3322/caac.21349
4. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928
5. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346–352. doi:10.1016/j.bbrc.2018.09.073
6. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38:98. doi:10.1186/s13046-019-1041-2
7. Xu JZ, CC S, XJ W, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi:10.1038/s41419-019-1300-3
8. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi:10.1186/s12943-019-1010-6
9. Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18:25. doi:10.1186/s12943-019-1010-6
10. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–1041. doi:10.2147/CMAR.S186857
11. Wei J, Wei W, Xu H, et al. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2019;1–7. doi:10.3233/CBM-182029
12. Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234:7247–7256. doi:10.1002/jcp.27482
13. Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;504:184–189. doi:10.1016/j.bbrc.2018.08.152
14. Guo JN, Li J, Zhu C-L, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi:10.2147/OTT.S123220
15. Ding X, Yang Y, Sun Y, Xu W, Su B, Zhou X. MicroRNA-585 acts as a tumor suppressor in non-small-cell lung cancer by targeting hSMG-1. Clin Transl Oncol. 2017;19:546–552. doi:10.1007/s12094-016-1562-5
16. Hu L, Wu H, Wan X, et al. MicroRNA-585 suppresses tumor proliferation and migration in gastric cancer by directly targeting MAPK1. Biochem Biophys Res Commun. 2018;499:52–58. doi:10.1016/j.bbrc.2018.03.116
17. Xie D, Liang Y, Su Y, An Y, Qu P. miR-152 inhibits proliferation of human endometrial cancer cells via inducing G2/M phase arrest by suppressing CDC25B expression. Biomed Pharmacother. 2018;99:299–305. doi:10.1016/j.biopha.2018.01.046
18. Lyons J, Bastian BC, McCormick F. MC1R and cAMP signaling inhibit cdc25B activity and delay cell cycle progression in melanoma cells. Proc Natl Acad Sci U S A. 2013;110:13845–13850. doi:10.1073/pnas.1201917110
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.