Back to Journals » Cancer Management and Research » Volume 12
Circular RNA hsa_circ_0003496 Contributes to Tumorigenesis and Chemoresistance in Osteosarcoma Through Targeting (microRNA) miR-370/Krüppel-Like Factor 12 Axis
Authors Xie C, Liang G, Xu Y, Lin E
Received 14 March 2020
Accepted for publication 24 July 2020
Published 9 September 2020 Volume 2020:12 Pages 8229—8240
DOI https://doi.org/10.2147/CMAR.S253969
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Chaofan Xie,1,* Guanzhao Liang,2,* Yinfeng Xu,1 Erhu Lin3
1Department of Orthopaedics, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, People’s Republic of China; 2Department of Emergency, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province, People’s Republic of China; 3Department of Spine Surgery, Shenzhen People’s Hospital, Shenzhen, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Erhu Lin Department of Spine Surgery
Shenzhen People’s Hospital, 1017 Dongmen North Road, Luohu District, Shenzhen, Guangdong Province 518020, People’s Republic of China
Tel +86-13760341589
Email [email protected]
Background: Osteosarcoma (OS) is the most common primary malignancy of bone with a high incidence in children. Circular RNAs (circRNAs) play crucial roles in the carcinogenesis and chemoresistance of OS. In the current work, we focused on the function and mechanism of hsa_circ_0003496 (circ_0003496) in OS progression and chemoresistance.
Materials and Methods: The expression levels of circ_0003496, miR-370 and Krüppel-like factor 12 (KLF12) mRNA were measured by quantitative real-time polymerase chain reaction (qRT-PCR). The Cell Counting Kit-8 (CCK-8) assay was used to assess the 50% inhibitory concentration (IC50) value and cell proliferation. Cell migration, invasion and apoptosis were detected by transwell assay and flow cytometry, respectively. Western blot analysis was performed to assess the protein level. Targeted relationships among circ_0003496, miR-370 and KLF12 were validated by dual-luciferase reporter, RNA immunoprecipitation (RIP) and RNA pull-down assays. Animal studies were carried out to observe the role of circ_0003496 in vivo.
Results: Our results indicated that circ_0003496 up-regulation was associated with doxorubicin (DXR) resistance of OS. Circ_0003496 knockdown repressed DXR-resistant OS cell proliferation, migration and invasion, and enhanced apoptosis and DXR sensitivity. Circ_0003496 functioned as a sponge of miR-370, and miR-370 mediated the regulatory effect of circ_0003496 depletion on DXR-resistant OS cell progression and DXR sensitivity. KLF12 was a direct target of miR-370, and miR-370 overexpression suppressed cell progression and enhanced DXR sensitivity by KLF12. Moreover, circ_0003496 protected against KLF12 repression through sponging miR-370. Additionally, circ_0003496 knockdown hampered tumor growth and promoted DXR sensitivity in vivo.
Conclusion: Our present work suggested that the knockdown of circ_0003496 suppressed OS progression and enhanced DXR sensitivity at least partially through modulating KLF12 expression via functioning as a miR-370 sponge, highlighting new opportunities for OS management.
Keywords: osteosarcoma, OS, circ_0003496, miR-370, Krüppel-like factor 12, KLF12, chemoresistance
Introduction
Osteosarcoma (OS) is the most common bone malignancy in children and young adults.1 In recent years, chemotherapy has significantly improved the long-term survival rates for OS patients.2 Nevertheless, the development of chemoresistance is still a major obstacle for OS management.3,4 Hence, a clearer understanding for what drives OS progression and chemoresistance is imperative to develop more effective strategies for OS therapy.
Circular RNAs (circRNAs) are endogenous covalently closed RNA molecules that exert critical functions in cancer biology.5,6 Recently, circRNAs have been implicated in the carcinogenesis and chemoresistance of OS.7,8 For instance, Chen et al reported that circRNA myosin-X contributed to OS progression through targeting microRNA (miRNA)-370-3p/RuvB-like protein 1 axis.9 Kun and colleagues uncovered that circRNA plasmacytoma variant translocation 1 promoted OS cell doxorubicin (DXR) and cisplatin resistance via the regulation of multidrug resistance polypeptide 1.10 Using high-throughput human circRNA microarray, hsa_circ_0003496 (circ_0003496) was found to be up-regulated in OS tissues.11 However, the function and mechanism of circ_0003496 in OS remain undefined.
MiRNAs, a class of small non-coding regions in RNA molecules of 19–23 nucleotides, have been identified to silence the expression of various oncogenes or tumor-suppressive genes during OS tumorigenic processes.12 Previous reports demonstrated that miR-370 was prominently down-regulated in OS and suppressed OS progression through targeting Forkhead box protein M1 or activating Wnt/β-catenin signaling pathway.13,14 CircRNAs were also underscored to exert essential functions via the gene-regulatory potential in OS pathogenesis through acting as efficient miRNA sponges.15,16 Using the online database, two complementary sequences among circ_0003496, miR-370 and Krüppel-like factor 12 (KLF12) were predicted, which promoted us to examine the miR-370/KLF12 axis as a potential mediator of circ_0003496 in regulating OS progression and chemoresistance.
In the present work, we firstly demonstrated that the reduced expression of circ_0003496 was associated with OS DXR resistance. Subsequently, we explored the function and mechanism of circ_0003496 on OS progression and chemoresistance in vitro and in vivo.
Materials and Methods
Clinical Tissues and Cells
In total, 35 primary OS patients and 35 recurrent patients after treatment with DXR-based on chemotherapy who underwent surgery at The First Affiliated Hospital of Sun Yat-sen University were enrolled in the current study. The clinicopathologic features of these patients are provided in Table 1. Fresh specimens were collected from these patients with prior written informed consent. All specimens were stored at −80°C. All protocols in the study were approved by the Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University.
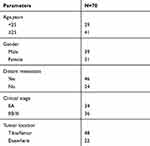 |
Table 1 The Clinicopathologic Features in Osteosarcoma Patients |
Human KHOS (ATCC®CRL-1544, American Type Culture Collection, ATCC, Manassas, VA, USA) and MG63 (ATCC®CRL-1427, ATCC) OS cell lines were maintained in a humidified atmosphere containing 5% CO2 in Eagle’s Minimum Essential Medium (ATCC) plus 10% fetal bovine serum (Lonza, Basel, Switzerland) at 37°C. Two DXR-resistant OS cell lines (KHOS/DXR and MG63/DXR cells) were established by 100Biotech (Hangzhou, China). To maintain the DXR resistance phenotype, additional 1 µg/mL of DXR (Sigma-Aldrich, Toyko, Japan) was added in the growth media.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was obtained from frozen tissues and cells using Trizol reagent (TaKaRa, Dalian, China) based on the guidance of the manufacturers. Circ_0003496 and KLF12 levels were determined using a TaKaRa Reverse Transcription Kit and the SYBR Green PCR Kit (TaKaRa), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization. The level of miR-370 was measured using TaqMan Reverse Transcription Kit and MicroRNA Assay Kit (all from Applied Biosystems, Courtaboeuf, France), and U6 was used as the internal control. The primers for PCR amplification were listed below: circ_0003496 (forward): 5ʹ-CCAACATCCCAGTCCTCAGT-3ʹ and circ_0003496 (reverse): 5ʹ-CCTGCTGTTGGGAAGTTTCA-3ʹ, KLF12 (forward): 5ʹ-TGGCAAAGCACAAATGGAC-3ʹ and KLF12 (reverse): 5ʹ-CTAAATGGTGAAATTGAACAAGG-3ʹ, GAPDH (forward): 5ʹ-GGGAGCCAAAAGGGTCAT-3ʹ and GADPH (reverse): 5ʹ-GAGTCCTTCCACGATACCAA-3ʹ, miR-370 (forward): 5ʹ-GAAGGATCCGTGTGCAAGGCGGGCTACTTG-3ʹ and miR-370 (reverse): 5ʹ-GGAGAATTCGATAGTTGCAGTGGGGGGTCCC-3ʹ, U6 (forward): 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and U6 (reverse): 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Determination of the 50% Inhibitory Concentration (IC50) Value, Cell Proliferation and Apoptosis
In the IC50 value studies, cells were exposed to various concentrations (0, 1, 2, 4, 8, 16, 32, 64 and 128 µg/mL) of DXR for 48 h. In the proliferation experiments, KHOS/DXR and MG63/DXR cells were performed the indicated transfection and then cultured for 24, 48 and 72 h. Then, cell viability was determined using the Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) based on the recommendations of the manufacturers. Absorbance was tested by a microplate reader set up at 450 nm. For the detection of cell apoptosis, transfected cells were stained with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) as per the manufacturing protocols (BD Biosciences, Heidelberg, Germany), and then the apoptotic cells were analyzed by flow cytometry.17
Cell Transfection and Lentiviral Vector Transduction
For loss-of-function experiments, KHOS/DXR and MG63/DXR cells were transfected with 50 nM of small interfering RNA (siRNA) against circ_0003496 (si-circ_0003496, GenePharma, Shanghai, China), commercial miR-370 inhibitor (in-miR-370, GenePharma) or a corresponding negative sequence (si-NC or in-miR-NC, GenePharma). For gain-of-function studies, 50 ng of pcDNA-based circ_0003496 and KLF12 overexpression plasmids (circ_0003496 and KLF12, VectorBuilder, Guangzhou, China) were transfected into cells, respectively, and a nontarget pcDNA vector (VectorBuilder) was used as the negative control. MiR-370 up-regulation was performed using 50 nM of synthetic miR-370 mimic (GenePharma), with a scrambled oligonucleotide sequence (miR-NC mimic, GenePharma) as the negative control. The commercially lipid-based Lipofectamine 2000 reagent (Invitrogen, Paisley, UK) was used for each transient transfection based on the suggestion of manufacturers. For in vivo studies, circ_0003496 depletion was achieved by corresponding lentiviral transduction. Lentiviral vectors harboring short hairpin RNA (shRNA) against circ_0003496 (sh-circ_0003496) or negative control (sh-NC) were constructed by Fulengen (Guangzhou, China) and used based on the manufactures’ instructions. Twenty-four hours later, the cells with positive transduction were selected with 10 μg/mL of puromycin (Sigma-Aldrich).
Transwell Migration and Invasion Assay
Transfected cells in 1% serum media were placed at 1.0 × 104 cells each well into 24-transwell inserts (8 µm pore size, Corning, Wiesbaden, Germany) for migration assays or at 2.0 × 104 cells each well into matrigel-precoated invasion chambers (Corning) for invasion assays. The lower chamber was added with 500 µL of 10% serum medium. Twenty-four hours later, the migrated or invaded cells were stained with 1% crystal violet and visualized under a microscope (Olympus, Tokyo, Japan) at 100× magnification. Images of 5 random fields were obtained and the average number of migrated or invaded cells was analyzed using Metamorph analysis software (Molecular Devices, San Jose, CA, USA).
Western Blot
Western blot analysis was carried out as described previously.18 Cell lysates were prepared using ice-cold lysis buffer. Total protein was separated by 8% SDS polyacrylamide gel electrophoresis and then transferred into nitrocellulose membranes (Millipore, Shanghai, China). The following antibodies were used: anti-multidrug resistance associated protein 1 (anti-MRP1, ab233383), anti-P-glycoprotein (anti-P-gp, ab216656), anti-lung cancer-related protein (anti-LRP, ab92544), anti-KLF12 (ab129459), anti-β-actin (ab8227) and horseradish peroxidase-conjugated IgG secondary antibody (ab150077, all from Abcam, Cambridge, UK).
Bioinformatics and Dual-Luciferase Reporter Assay
The targeted miRNAs of circ_0003496 and miR-370 molecular targets were predicted using the online database Circinteractome (https://circinteractome.nia.nih.gov/miRNA_Target_Sites/mirna_target_sites.html) and TargetScan v.7.1 (http://www.targetscan.org/vert_71/?tdsourcetag=s_pcqq_aiomsg), respectively. The segmental sequences of circ_0003496 harboring the miR-370-binding sites and the full-length of KLF12 3ʹ-untranslated region (3ʹ-UTR) were inserted into the pmirGLO vector (Promega, Southampton, UK). Site-directed mutations in the target region were constructed using the TaKaRa MutanBEST Kit following the protocols of the manufacturers. KHOS/DXR and MG63/DXR cells were transfected with 10 ng of each reporter construct and miR-370 mimic or miR-NC mimic for 48 h, followed by the detection of luciferase activities using the Promega Dual-luciferase Reporter Assay System.
RNA Immunoprecipitation (RIP) and RNA Pull-Down Assays
In RIP assays, KHOS/DXR and MG63/DXR cells were transfected with miR-370 mimic or miR-NC mimic, lysed in lysis buffer, and then incubated with anti-Argonaute2 (anti-Ago2, ab57113, Abcam) or anti-IgG antibody at 4°C for 2 h before adding the protein A/G beads (Pierce, Bonn, Germany) for 4 h. In RNA pull-down assays, cell lysates were incubated with Biotin-labeled circ_0003496 (Bio-circ_0003496 WT), mutated circ_0003496 (Bio-circ_0003496 MUT) in the miR-370-binding sites or a negative control (Bio-NC, all from ACROBiosystems, Beijing, China) at 4°C for 4 h, and then incubated with Streptavidin agarose beads (Pierce) for 2 h. In both assays, beads were collected, and total RNA was isolated to measure circ_0003496 and miR-370 levels by qRT-PCR.
Animal Studies
All animal experiments were performed in compliance with the National Standard of the Care and Use of Laboratory Animals, following the approved protocol by the Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University. In the animal work, 5-week-old BALC/c nude mice (n = 24, Cyagen, Suzhou, China) were used. MG63/DXR cells (5.0 × 106) stably transduced with sh-circ_0003496 or sh-NC were subcutaneously inoculated into nude mice, followed by the administration with DXR (5 mg/kg) or PBS through tail vein injection every other days after 5 days first implantation. On days 7, 14, 21 and 28 after cell inoculation, tumor volume was measured and calculated using the equation: V = Length (mm) × Width (mm)2/2. In the end of the experiments, all mice were sacrificed, and tumor tissues were excised.
Statistical Analysis
All data were presented as mean ± standard deviation. Statistical significances were analyzed by a tailed Student’s t-test or one-way analysis of variance (ANOVA). Differences between the expression levels of circ_0003496, miR-370 and KLF12 mRNA in OS tissues were compared by the Mann–Whitney test. P values at 0.05 or smaller were considered significant.
Results
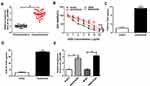
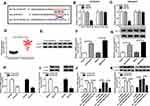
Up-Regulation of circ_0003496 Was Associated with DXR Resistance of OS
To preliminarily observe the involvement of circ_0003496 in OS chemoresistance, we firstly assessed its expression in OS tissues from primary patients (Chemosensitive tissues) and recurrent patients (Chemoresistant tissues) using qRT-PCR. As shown in Figure 1A, circ_0003496 level was higher in the Chemoresistant group than that in the Chemosensitive group. Then, we established two DXR-resistant OS cell lines (KHOS/DXR and MG63/DXR). The data of CCK-8 assays showed that the IC50 values of DXR-resistant OS cells were significantly elevated when comparing to their parents (Figure 1B–D). Moreover, in contrast to the DXR-sensitive OS cells, circ_0003496 expression was prominently up-regulated in the DXR-resistant OS cells (Figure 1E).
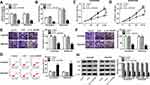
Circ_0003496 Regulated Cell Proliferation, Migration, Invasion, Apoptosis and DXR Sensitivity in DXR-Resistant OS Cells
To determine the functional role of circ_0003496 in OS, we performed loss-of-function analyses in DXR-resistant OS cells using siRNA against circ_0003496 (si-circ_0003496). As shown by qRT-PCR, in comparison to the negative control, si-circ_0003496 transfection led to a remarkable reduction in the level of circ_0003496 (Figure 2A). CCK-8 assays revealed that circ_0003496 knockdown significantly attenuated the IC50 value for DXR (Figure 2B) and cell proliferation (Figure 2C and D). Moreover, circ_0003496 knockdown strikingly repressed cell migration and invasion (Figure 2E and F) and promoted cell apoptosis (Figure 2G). Furthermore, the results of Western blot showed that circ_0003496 knockdown resulted in decreased expression levels of the related multidrug resistance (MDR) proteins (MRP1, P-gp and LRP) in both KHOS/DXR and MG63/DXR cells (Figure 2H). Additionally, in contrast to the control group, the elevated expression of circ_0003496 significantly increased the IC50 value for DXR, enhanced cell proliferation, migration and invasion, and repressed cell apoptosis, as well as elevated the levels of MRP1, P-gp and LRP in both KHOS/DXR and MG63/DXR cells (Supplementary Figure 1).
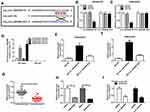
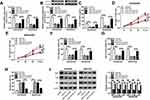
Circ_0003496 Acted as a Molecular Sponge of miR-370
To further understand the mechanism of circ_0003496 in OS, we used online database Circinteractome to predict the potential miRNAs that bind to circ_0003496. These predicted data revealed a putative binding sequence for miR-370 within circ_0003496 (Figure 3A). To validate this, we carried out dual-luciferase reporter, RIP and RNA pull-down assays. In contrast, the transfection of miR-370 mimic significantly down-regulated the luciferase activity of wild-type reporter construct in both KHOS/DXR and MG63/DXR cells (Figure 3B and C). When the target sequence was mutated, little change was observed in luciferase in the presence of miR-370 mimic (Figure 3B and C). The data of RIP experiments revealed that the enrichment level of circ_0003496 was drastically elevated by miR-370 mimic using anti-Ago2 antibody (Figure 3D). Moreover, RNA pull-down results demonstrated that miR-370 was highly enriched by wild-type Bio-circ_0003496 (Figure 3E and F). When the mutant-type Bio-circ_0003496 in the target sequence was tested, the mutation no longer elicited such effect (Figure 3E and F), indicating the validity of the binding sequence for interaction. The data of qRT-PCR also showed a significant down-regulation of miR-370 expression in the Chemoresistant OS tissues and cells compared to the Chemosensitive controls (Figure 3G and H). Furthermore, circ_0003496 overexpression prominently decreased the level of miR-370 in the two OS/DXR cells (Figure 3I).
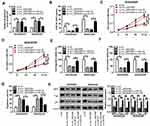
Circ_0003496 Knockdown Regulated Cell Proliferation, Migration, Invasion, Apoptosis and DXR Sensitivity in DXR-Resistant OS Cells by Up-Regulating miR-370
Then, we determined whether miR-370 acted as a key mediator of circ_0003496 in regulating DXR-resistant OS cell progression and DXR sensitivity. In comparison to the negative control, in-miR-370 transfection significantly reversed the increase of si-circ_0003496 on miR-370 level in both KHOS/DXR and MG63/DXR cells (Figure 4A). Moreover, the restored miR-370 expression drastically abrogated circ_0003496 depletion-mediated IC50 value diminishment (Figure 4B), cell proliferation inhibition (Figure 4C and D), cell migration and invasion repression (Figure 4E and F) and cell apoptosis enhancement (Figure 4G), as well as MDR proteins (MRP1, P-gp and LRP) reduction (Figure 4H).
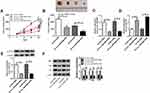
Circ_0003496 Regulated KLF12 Expression by Sponging miR-370
Next, we performed a detailed analysis for miR-370 targets using TargetScan v.7.1 software. Of interest, a putative complementary region for miR-370 was found within the 3ʹ-UTR of KLF12 (Figure 5A). To verify this, we cloned KLF12 3ʹ-UTR into a luciferase vector and mutated the target sequence. With KLF12 3ʹ-UTR wild-type reporter and miR-370 overexpression caused a striking reduction in luciferase activity (Figure 5B and C). However, little change was observed in the luciferase activity of mutant-type reporter in the presence of miR-370 mimic (Figure 5B and C). qRT-PCR results also revealed a significant up-regulation of KLF12 level in the Chemoresistant OS tissues and cells when comparing to the Chemosensitive controls (Figure 5D–G). Moreover, the elevated level of miR-370 led to a striking reduction in KLF12 expression at both mRNA and protein levels in both KHOS/DXR and MG63/DXR cells (Figure 5H and I). Additionally, we further validated whether circ_0003496 modulated KLF12 expression in the two DXR-resistant OS cells. As expected, in contrast to their counterparts, circ_0003496 up-regulation significantly elevated KLF12 expression at both mRNA and protein levels, and this effect was remarkably abrogated by miR-370 mimic (Figure 5J and K).
KLF12 Was a Functional Target of miR-370 in Regulating Cell Proliferation, Migration, Invasion, Apoptosis and DXR Sensitivity in DXR-Resistant OS Cells
To provide further mechanistic insight into the link between miR-370 and KLF12 on DXR-resistant OS cell progression and DXR sensitivity, KHOS/DXR and MG63/DXR cells were transfected with miR-370 mimic alone or together with KLF12 overexpression plasmid. By contrast, the transfection of KLF12 overexpression plasmid significantly abolished the reduction of miR-370 overexpression on KLF12 expression (Figure 6A and B). Moreover, miR-370 overexpression led to a significant decline in the IC50 value (Figure 6C), a striking repression in cell proliferation (Figure 6D and E), and a distinct suppression in cell migration and invasion (Figure 6F and G), as well as a prominent promotion in cell apoptosis (Figure 6H). Moreover, the up-regulation of miR-370 resulted in reduced MDR proteins (MRP1, P-gp and LRP) expression (Figure 6I). Nevertheless, KLF12 expression restoration drastically reversed these effects of miR-370 overexpression in the two OS/DXR cells (Figure 6C–I).
Circ_0003496 Knockdown Mitigated Tumor Growth and Promoted DXR Sensitivity in vivo
Lastly, we determined the effect of circ_0003496 on OS progression and DXR resistance in vivo. In comparison to their counterparts, sh-circ_0003496 transduction or DXR treatment led to a significant inhibition in tumor growth (Figure 7A and B). Simultaneous sh-circ_0003496 transduction and DXR treatment triggered a more strong suppression in tumor growth (Figure 7A and B). The data of qRT-PCR showed that the transduction of sh-circ_0003496 resulted in reduced circ_0003496 expression in xenograft tissues with or without DXR treatment (Figure 7C). Moreover, miR-370 was up-regulated and KLF12 was down-regulated in tumor tissues derived from the circ_0003496-silencing MG63/DXR cells (Figure 7D and E). Additionally, the levels of MRP1, P-gp and LRP were decreasing in the xenograft tumors derived from circ_0003496-silencing MG63/DXR cells (Figure 7F).
Discussion
CircRNAs are being found to have relevance to the carcinogenesis of OS, demonstrating their potential as biomarkers and targets for OS management.19 Moreover, circRNAs have been highlighted to be related to the development of OS chemoresistance.8,10 In the current study, we aimed to explore the biological role and molecular mechanism of circ_0003496 in OS progression and chemoresistance.
Previous research reported that circ_0003496 was up-regulated in vein samples of patients with primary great saphenous vein varicosities using the high-throughput sequencing.20 Similarly, circ_0003496 was highly expressed in OS tissues using high-throughput circRNA microarray.11 These findings described above provided the possibility that circ_0003496 might be involved in the progression and chemoresistance of OS. To verify this, we firstly validated circ_0003496 expression in OS tissues, and our data demonstrated a significant up-regulation of circ_0003496 in the Chemoresistant OS tissues and cells. MDR proteins (MRP1, P-gp and LRP) are important for the development of drug resistance in OS.21 In this work, we were first to uncover that circ_0003496 knockdown hampered DXR-resistant OS cell proliferation, migration and invasion and promoted cell apoptosis and DXR sensitivity in vitro. Moreover, our data suggested that circ_0003496 depletion hindered tumor growth and promoted DXR sensitivity in vivo.
Then, we used online database Circinteractome to help identify the targeted miRNAs of circ_0003496. Among the nineteen targets, miR-370 was fascinating in the current project, considering its crucial roles in a wide range of human tumors, such as ovarian cancer, gastric cancer and bladder cancer.22–24 MiR-370 was also reported to regulate tumor cell chemoresistance via targeting genes in ovarian cancer and chronic myeloid leukemia.25,26 Moreover, miR-370 overexpression repressed OS cell proliferation and colony formation via activating Wnt/β-catenin signaling pathway.14 In the present work, we firstly validated that circ_0003496 acted as a miR-370 sponge in DXR-resistant OS cells. Our data also demonstrated that the increased level of miR-370 suppressed OS cell progression and enhanced DXR sensitivity, consistent with an earlier document.13 Furthermore, we firstly uncovered the regulation of circ_0003496 on DXR-resistant OS cell progression and DXR sensitivity by miR-370.
Here, we confirmed that KLF12 was a direct target of miR-370. KLF12, a member of Krüppel-like factors that are DNA-binding transcriptional regulators, was identified to function as vital regulators in human cancers, such as colorectal cancer, nasopharyngeal carcinoma and gastric cancer.27–29 Moreover, the up-regulation of KLF12 was demonstrated to contribute to OS cell growth and chemoresistance in vitro and in vivo.30 In the present work, we were first to highlight that KLF12 was a functionally important target of miR-370 in regulating OS cell progression and enhanced DXR sensitivity. Similarly, Xu et al showed that miR-382 suppressed OS tumorigenesis and promoted chemosensitivity through targeting KLF12.30 More interestingly, our data for the first time substantiated that circ_0003496 controlled KLF12 expression through acting as a miR-370 sponge in DXR-resistant OS cells. Additionally, in vivo assay demonstrated that circ_0003496 knockdown resulted in increased miR-370 expression and decreased KLF12 level in xenograft tissues. Therefore, more in vivo investigations about the correlation between circ_0003496 and the miR-370/KLF12 axis in OS progression and chemoresistance will be conducted in further work. Additionally, osteoblasts, the bone-forming cells, can influence whole-body metabolism by responding to metabolic hormones and producing endocrine factors.31 Hu et al reported that miR-132-3p negatively regulated osteogenic differentiation and osteogenesis.32 Cao et al uncovered that long noncoding RNA Linc02349 enhanced osteogenesis of human umbilical cord-derived from stem cells by targeting miR-25-3p and miR-33-5p.33 The evidence illuminated the involvement of noncoding RNAs in osteogenesis and bone-related diseases. A further challenge is to explore whether the novel mechanism, the circ_0003496/miR-370/KLF12 axis, is involved in the osteogenesis and other bone diseases.
In conclusion, the current work identified that the knockdown of circ_0003496 repressed OS progression and enhanced DXR sensitivity in vitro and in vivo possibly through protecting against KLF12 repression via sponging miR-370. Thus, circ_0003496 and its molecular mediators might be novel therapeutic opportunities for OS treatment.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
1. Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheum Ther. 2017;4(1):25–43. doi:10.1007/s40744-016-0050-2
2. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. doi:10.1080/14737140.2018.1413939
3. Rejniak KA, Lloyd MC, Reed DR, Bui MM. Diagnostic assessment of osteosarcoma chemoresistance based on virtual clinical trials. Med Hypotheses. 2015;85(3):348–354. doi:10.1016/j.mehy.2015.06.015
4. Kim M, Kim DJ. GFRA1: a novel molecular target for the prevention of osteosarcoma chemoresistance. Int J Mol Sci. 2018;19(4):1078. doi:10.3390/ijms19041078
5. Su M, Xiao Y, Ma J, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18(1):90. doi:10.1186/s12943-019-1002-6
6. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
7. Liu W, Zhang J, Zou C, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43(3):969–985. doi:10.1159/000481650
8. Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin Z, Jian-Ping H, Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10(10):1327–1346. doi:10.2217/epi-2018-0023
9. Chen J, Liu G, Wu Y, et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18(1):150. doi:10.1186/s12943-019-1076-1
10. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321–330. doi:10.7150/ijbs.24360
11. Liu X, Zhong Y, Li J, Shan A. Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget. 2017;8(70):114829–114838. doi:10.18632/oncotarget.22162
12. Kushlinskii NE, Fridman MV, Braga EA. Molecular mechanisms and microRNAs in osteosarcoma pathogenesis. Biochemistry (Mosc). 2016;81(4):315–328. doi:10.1134/S0006297916040027
13. Duan N, Hu X, Yang X, Cheng H, Zhang W. MicroRNA-370 directly targets FOXM1 to inhibit cell growth and metastasis in osteosarcoma cells. Int J Clin Exp Pathol. 2015;8(9):10250–10260.
14. Zhang W, Duan N, Zhang Q, et al. DNA methylation mediated down-regulation of miR-370 regulates cell growth through activation of the Wnt/β-catenin signaling pathway in human osteosarcoma cells. Int J Biol Sci. 2017;13(5):561–573. doi:10.7150/ijbs.19032
15. Song Y-Z, Li J-F. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495(3):2369–2375. doi:10.1016/j.bbrc.2017.12.050
16. Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. 2018;502(4):465–471. doi:10.1016/j.bbrc.2018.05.184
17. Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109:226–232. doi:10.1016/j.biopha.2018.10.072
18. Yan P, Su Z, Zhang Z, Gao T. LncRNA NEAT1 enhances the resistance of anaplastic thyroid carcinoma cells to cisplatin by sponging miR‑9‑5p and regulating SPAG9 expression. Int J Oncol. 2019;55(5):988–1002. doi:10.3892/ijo.2019.4868
19. Wang C, Jing J, Cheng L. Emerging roles of non-coding RNAs in the pathogenesis, diagnosis and prognosis of osteosarcoma. Invest New Drugs. 2018;36(6):1116–1132. doi:10.1007/s10637-018-0624-7
20. Zhang W, Li L, Si Y, et al. Identification of aberrant circular RNA expression and its potential clinical value in primary great saphenous vein varicosities. Biochem Biophys Res Commun. 2018;499(2):328–337. doi:10.1016/j.bbrc.2018.03.156
21. Li S, Sun W, Wang H, Zuo D, Hua Y, Cai Z. Research progress on the multidrug resistance mechanisms of osteosarcoma chemotherapy and reversal. Tumour Biol. 2015;36(3):1329–1338. doi:10.1007/s13277-015-3181-0
22. Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi:10.1016/j.omtn.2018.08.010
23. Ning T, Zhang H, Wang X, et al. miR-370 regulates cell proliferation and migration by targeting EGFR in gastric cancer. Oncol Rep. 2017;38(1):384–392. doi:10.3892/or.2017.5660
24. Huang X, Zhu H, Gao Z, et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J Biol Chem. 2018;293(18):6693–6706. doi:10.1074/jbc.RA118.001689
25. Chen X-P, Chen Y-G, Lan J-Y, Shen Z-J. MicroRNA-370 suppresses proliferation and promotes endometrioid ovarian cancer chemosensitivity to cDDP by negatively regulating ENG. Cancer Lett. 2014;353(2):201–210. doi:10.1016/j.canlet.2014.07.026
26. Zhou M, Zeng J, Wang X, et al. MiR-370 sensitizes chronic myeloid leukemia K562 cells to homoharringtonine by targeting Forkhead box M1. J Transl Med. 2013;11:265. doi:10.1186/1479-5876-11-265
27. Kim S-H, Park -Y-Y, Cho S-N, Margalit O, Wang D, DuBois RN. Krüppel-like factor 12 promotes colorectal cancer growth through early growth response protein 1. PLoS One. 2016;11(7):e0159899–e0159899. doi:10.1371/journal.pone.0159899
28. Song P, Yin S-C. Long non-coding RNA 319 facilitates nasopharyngeal carcinoma carcinogenesis through regulation of miR-1207-5p/KLF12 axis. Gene. 2019;680:51–58. doi:10.1016/j.gene.2018.09.032
29. Nakamura Y, Migita T, Hosoda F, et al. Krüppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125(8):1859–1867. doi:10.1002/ijc.24538
30. Xu M, Jin H, Xu C-X, et al. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5(19):9472–9483. doi:10.18632/oncotarget.2418
31. Dirckx N, Moorer MC, Clemens TL, et al. The role of osteoblasts in energy homeostasis. Nat Rev Endocrinol. 2019;15(11):651–665. doi:10.1038/s41574-019-0246-y
32. Hu Z, Zhang L, Wang H, et al. Targeted silencing of miRNA-132-3p expression rescues disuse osteopenia by promoting mesenchymal stem cell osteogenic differentiation and osteogenesis in mice. Stem Cell Res Ther. 2020;11(1):58. doi:10.1186/s13287-020-1581-6
33. Cao L, Liu W, Zhong Y, et al. Linc02349 promotes osteogenesis of human umbilical cord-derived stem cells by acting as a competing endogenous RNA for miR-25-3p and miR-33b-5p. Cell Prolif. 2020;53(5):e12814. doi:10.1111/cpr.12814
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.