Back to Journals » Journal of Inflammation Research » Volume 15
Circular RNA Fbxl5 Regulates Cardiomyocyte Apoptosis During Ischemia Reperfusion Injury via Sponging microRNA-146a
Authors Li D, You J, Mao C, Zhou E, Han Z, Zhang J, Zhang T, Wang C
Received 3 February 2022
Accepted for publication 13 April 2022
Published 21 April 2022 Volume 2022:15 Pages 2539—2550
DOI https://doi.org/10.2147/JIR.S360129
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Monika Sharma
Dongjiu Li,1,* Jiayin You,2,* Chengyu Mao,1,* En Zhou,1,* Zhihua Han,1 Junfeng Zhang,1 Tiantian Zhang,1 Changqian Wang1
1Department of Cardiology, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200011, People’s Republic of China; 2Department of Emergency, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200011, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tiantian Zhang; Changqian Wang, Email [email protected]; [email protected]
Objective: Cardiomyocyte apoptosis critically contributes to ischemia reperfusion injury (IRI), which lacks effective therapeutic strategies. Circular RNAs (circRNAs) serve as novel diagnostic and therapeutic targets in various cardiovascular diseases. CircRNA Fbxl5 is one of the abundantly expressed circRNAs in the heart and its role in myocardial IRI remains elusive.
Materials and Methods: Wild-type (WT) mice and neonatal mice ventricular myocytes (NMVMs) were used and subjected to myocardial IRI and anoxia reoxygenation (AR), respectively. Molecular and histological analyses and echocardiography were used to determine the extent of apoptosis, infarct size, and cardiac function.
Results: We found that circRNA Fbxl5 was significantly upregulated in the myocardium, as well as in NMVMs subjected to AR. Knockdown of circRNA Fbxl5 ameliorated cardiomyocyte apoptosis, thereby decreasing infarct size and preserving cardiac function. Additionally, in vitro knockdown of circRNA Fbxl5 in NMVMs subjected to AR recapitulated the in vivo findings. Mechanistically, we identified that circRNA Fbxl5 directly sponged and suppressed the endogenous microRNA-146a (miR-146a), thereby weakening its inhibitory effect on MED1, which could further promote the apoptosis of cardiomyocytes.
Conclusion: Our findings revealed a novel and critical role for circRNA Fbxl5 in regulating cardiomyocyte apoptosis, and added additional insight into circRNAs mediated during myocardial IRI. The underlying miR-146a-MED1 signaling serves as an important cascade in regulating the apoptosis of cardiomyocytes.
Keywords: myocardial ischemia reperfusion injury, circular RNA, microRNA-146a, cardiomyocyte apoptosis
Introduction
Acute myocardial infarction is the leading cause of death worldwide. Timely myocardial reperfusion is the top priority for limiting infarct size and preserving cardiac functions.1 However, reperfusion per se can further lead to cardiomyocyte death, termed as ischemia reperfusion injury (IRI).2 At present, there is no effective therapeutic strategy for myocardial IRI.3 Therefore, revealing novel molecular mechanisms underlying IRI is of great significance.
Circular RNAs (CircRNAs) represent one of the widely expressed noncoding RNAs that form covalently closed continuous loops.4 In contrast to previous viewpoint that circRNAs are merely by-product of eukaryotic transcriptomes, recent studies have shown that some circRNAs regulate gene expression mainly through acting as endogenous sponge RNAs to interact with microRNAs (miRNAs) and influence the expressions of miRNA target genes.5 A previous study demonstrated that circRNA ciRS-7 acts as a miRNA-7 sponge to suppress miRNA-7 activity, resulting in the increased levels of miRNA-7 targets.6 Another study showed that circRNA CDR1as functions to bind miRNA-7 in neuronal tissues and sequesters away the miRNA-7 from its target sites.7 In addition, dysregulated circRNAs were associated with cardiovascular diseases, including heart failure,8 myocardial IRI9,10 and myocardial infarction and ventricular remodeling,11,12 indicating that they play important roles under these pathological conditions.
Our previous study demonstrated that miR-146a was protective against myocardial IRI and attenuated cardiomyocyte apoptosis via directly targeting MED1, which might mediate the apoptosis through regulating p53 signaling pathway.13 In the present study, we identified circRNA Fbxl5, one of the abundantly expressed circRNAs in the heart, which could function as miR-146a sponge. We aimed to investigate the role of circRNA Fbxl5 in myocardial IRI. Mechanistically, we attempted to indicate whether the miR-146a-MED1 cascade could contribute to the regulatory role of circRNA Fbxl5 in myocardial IRI.
Materials and Methods
Male adult C57BL/6 mice (age, 8–10-week-old) were purchased from Shanghai SLAC Experimental Animal Co., Ltd. (Shanghai, China). Mice were kept under standard housing conditions (temperature,at 21±1 °C; humidity, 55–60%) in the division of Laboratory Animal Resources of Shanghai Ninth People’s Hospital, according to the guidelines for the welfare of laboratory animals.14 Throughout the experiments, mice were fed with a standard chow diet and tap water. The animal room was cleaned regularly during the housing period. All animal experiments were approved by the Institutional Ethics Committee of the Shanghai Ninth People’s Hospital (Approval No. HKDL2017300), and were performed in accordance with the international ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines for animal experiments.15
Animal Model of Myocardial IRI
Myocardial IRI was surgically induced in mice as described previously.9 Briefly, for detecting the variation of miR-146a after myocardial IRI, mice were randomly divided into two groups, including sham and IRI groups, and the latter was further divided into four subgroups according to different durations of IRI (eg, 0.5, 1, 2, and 3 h; n=6 mice per group). All mice except for those in sham group were subjected to ischemia (45 min), followed by different durations of IRI. Mice in the sham group underwent the same procedure except that the snare was left untied. In order to further evaluate the effects of circRNA Fbxl5 on myocardial IRI, mice were divided into two groups, including sham and IR groups. Two weeks before surgery, mice in the sham group received AAV9 vectors (vehicle), while mice in the IR group received AAV9-TnT-si-circRNA (si-circ) or negative control (si-NC) or vehicle through tail vein. Mice in the IR group were subjected to ischemia (45 min), followed by IRI. Hearts were harvested for histological experiments and Western blotting 1 day after IRI, and echocardiography was performed 1 week after IRI.
After IRI, Evans blue/2,3,5-triphenyltetrazolium (TTC; Sigma-Aldrich, St. Louis, MO, USA) staining was performed as previously described.10 The infarct size, myocardial area at risk (AAR), and nonischemic left ventricle (LV) were assessed using the Image J 1.48 software.
Adeno-Associated Virus Serotype 9 (AAV9) Mediated Gene Delivery in the Heart
AAV9 expressing green fluorescent protein (GFP) alone or harboring siRNA targeting circRNA Fbxl5 (si-circ) or negative control (si-NC) could be synthesized and produced by Zorin Co., Ltd. (Shanghai, China) according to the manufacturer’s protocol. For the experiment protocol, mice were randomly divided into four groups, including sham (n = 6), IRI (n = 6), AAV9-si-circ (n = 6), and AAV9-NC (n = 6) groups. One hundred microliters of each AAV9 (2×1011 vector genomes) in phosphate-buffered saline (PBS) solution was loaded into a 1-mL syringe attached to a 29G needle. The AAV9 solution was injected into tail vein as described previously.16 The circRNA Fbxl5 knockdown in myocardium was identified after 2 weeks of injection, and thereafter myocardial IRI was induced. Mice in the sham and IRI groups received an equal volume of AAV9 vectors.
Echocardiography
Echocardiography was carried out one week after myocardial IRI using a 30-MHz ultrasound probe (Vevo3100, Box 66 Toronto, ON, Canada). Heart rate, left ventricular end-systolic diameter (LVESD), and left ventricular end-diastolic diameter (LVEDD) were measured. The left ventricular ejection fraction (LVEF) and fractional shortening (FS) were calculated as follows: LVEF (%) = [(LVEDD3-LVESD3)/LVEDD3] × 100%; FS (%) = [(LVEDD-LVESD)/LVEDD] × 100%. All parameters were measured by one experienced echocardiographer who was blinded to the study groups.
Histology
To evaluate cardiomyocyte apoptosis, the sections of myocardial tissues were stained with terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) using a kit (#11,684,795,910; Roche, Basel, Switzerland), according to the manufacturer’s instructions. At least four slides per heart tissue were chosen to evaluate the number and percentage of TUNEL-positive cells. For each slide, five randomly selected fields were selected and the cardiomyocytes per field were counted.
Isolation and Culture of Neonatal Mice Ventricular Myocytes (NMVMs) and Cardiac Fibroblasts (CFs)
NMVMs and CFs were isolated from neonatal mice that aged 1–2 days as described previously.17 Briefly, heart tissues of neonatal mice were quickly rinsed in 75% ethanol and decapitated. After dissection, ventricles were washed with PBS at 4 °C. The heart tissues were then digested with 0.25% trypsin at 37 °C. The cell suspension was collected and centrifuged at 200 g for 5 min, and the pellet was re-suspended in a Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), streptomycin (100 µg/mL), and penicillin (100 U/mL) at 37 °C under 5% CO2. The resuspension was pre-plated onto a culture flask at 37 °C for 90 min, in which CFs were attached to the bottom of the flask. The supernatant was collected and plated in 10 µg/mL laminin-coated culture dishes. CFs from passages 2–4 were used for subsequent experiments.
An in vitro AR Model for NMVMs and CFs
An in vitro AR model was established using an anoxic chamber pre-filled with saturated water content of gases, containing 5% CO2 and 95% nitrogen. NMVMs and CFs were cultured under anoxic conditions for 2 h, and then subjected to reoxygenation (95% O2 and 5% CO2) for 6 h. The negative control cells were cultured in a normoxic chamber (21% O2 and 5% CO2) supplemented with 10% FBS (Gibco, New York, NY, USA). TUNEL staining was performed by an in situ cell death detection kit (Roche, Basel, Switzerland) to evaluate cardiomyocyte apoptosis.
Dual-Luciferase Reporter Assay
Wild-type (WT) or mutated (MUT) circRNA Fbxl5 binding sequences and miR-146a mimics (including negative control) were constructed by Zorin Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions. Briefly, 293T cells were seeded into 24-well plates (density, 2×104 cells/well). Then, 293T cells were transfected with plasmid (500 ng) and miR-146a mimics or negative control (50 nM) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Cells were collected after 24 h for detecting firefly luciferase and Renilla luciferase activities using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
RNA Immunoprecipitation (RIP) Assay
A Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore, Burlington, MA, USA) was used for AGO2 protein-based RIP assay. Experimental process was performed according to the manufacturer’s instructions. Anti-AGO2 antibody was purchased from Abcam (Cambridge, UK; ab32381).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from NMVMs and CFs using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA concentration was quantified using NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). A Color Reverse Transcription kit (EZBioscience, Roseville, MN, USA) was utilized to perform the reverse transcription of cDNA. The expression level of circRNA Fbxl5 was measured using a PrimerScriptTM RT reagent kit (TaKaRa, Shiga, Japan; Cat. No. RR037A) using α-tubulin as an internal control. The forward primer sequence of circRNA Fbxl5 was GATGAGAAGGCAGATACAGAAGGA. The reverse primer sequence of circRNA Fbxl5 was TACTTGAGAACTTTCTGTCGCTCC. The miR-146a level was quantified by the Bulge-Loop mmu-miR-146a-5p Primer system (Cat. No. MQPS0002462-1) using U6 as an internal control. All qRT-PCR analyses were performed on an ABI 7500 System (Applied Biosystems, Waltham, MA, USA).
Eligibility Criteria for Predicting circRNAs
TargetScan and miRanda were used to predict circRNAs that most likely sponge miR-146a. Predictions were ranked based on the predicted efficacy of targeting as calculated using cumulative weighted context scores of the sites. We selected top 10 circRNAs that most likely sponge miR-146a for further validation, as shown in Table 1.
 |
Table 1 Top 10 circRNAs That Most Likely Sponge miR-146a |
Western Blotting
Western blotting was performed as previously described.18 In brief, total protein was extracted from frozen ventricular tissues, NMVMs and CFs using the RIPA lysis buffer (Beyotime, Shanghai, China). A total of 15 µg of protein was subjected to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and then, transferred onto 0.45-µm polyvinylidene difluoride (PVDF) membranes (Millipore). After that, the blots were blocked with a rapid blocking buffer (Shanghai EpiZyme Biotechnology Co., Ltd., Shanghai, China), followed by overnight incubation with primary antibodies at 4 °C. All primary antibodies were diluted (1:1000) in 5% bovine serum albumin (BSA). After thrice washing with TBST, the membranes were incubated for 1 h in secondary horseradish peroxidase (HRP)-conjugated antibodies (goat anti-rabbit IgG (H&L); Cat. No. ab81053; Abcam) diluted (1:5000) in 5% BSA. The blots were visualized using the ECL reagent kit (Millipore). ImageJ 1.48 software was used to quantify the intensity of the protein blots. The antibodies used in the current study are listed in Table 2.
 |
Table 2 Antibodies Used for Western Blotting |
Statistical Analysis
Data were expressed as mean ± standard deviation (SD) of at least three independent experiments. Continuous variables were compared between groups using two-tailed Student’s t-test or one-way analysis of variance (ANOVA), followed by the least significant difference t-test and Dunnett’s t-test via SPSS 19.0 software (IBM, Armonk, NY, USA). Normality and homogeneity of variance of all data were examined before making comparisons between groups. GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA) was used to carry out statistical analysis. P < 0.05 was considered statistically significant.
Results
IR Significantly Downregulated the Expression Level of miR-146a and Upregulated the Expression Level of circRNA Fbxl5
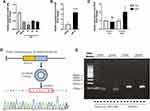
MiR-146a was demonstrated to protect the myocardium from IRI in our previous study.13 To further explore the variation of miR-146a after IR, we first examined its expression level at sham group and at 30 min, 1 h, 2 h, and 3 h after IRI in the mouse model. As shown in Figure 1A, it was revealed that the expression level of miR-146a in myocardium of mice significantly decreased after IR, and the lowest expression level was detected at 1 h after IR. In order to indicate whether circRNA functions as miR-146a sponge, we predicted the top 10 circRNAs that most likely sponge miR-146a using TargetScan and miRanda (Table 1). Then, we used qRT-PCR to detect these circRNAs after 1 h of IRI in the mouse model. Among these circRNAs, we identified circRNA Fbxl5, which was significantly upregulated at 1 h after IR (Figure 1B). Subsequently, we explored the expression level of circRNA Fbxl5 in NMVMs and CFs exposed to anoxia/reoxygenation (A/R). As displayed in Figure 1C, circRNA Fbxl5 was equally expressed in NMVMs and CFs, whereas it was significantly upregulated in NMVMs after A/R. Sanger sequencing of the amplified products indicated that circRNA Fbxl5 was produced from Fbxl5 gene and demonstrated the head-to-tail splicing of exon 4 (100-nt) (Figure 1D). Divergent primers only amplified circRNA Fbxl5 in cDNA, rather than genomic DNA (gDNA) (Figure 1E).
CircRNA Fbxl5 Acted as an Efficient miR-146a Sponge
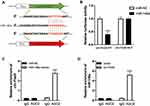
Given that circRNA Fbxl5 was abundantly expressed in cardiomyocytes and its expression level in ventricular myocardium after IRI was opposite to that of miR-146a, we investigated the ability of circRNA Fbxl5 to bind to miR-146a. Using miRBase, we found that miR-146a contained circRNA Fbxl5 binding site. As measured by the dual-luciferase reporter system, we noted that overexpression of miR-146a suppressed the activity of luciferase reporter encompassing circRNA Fbxl5 3’-UTR-WT rather than circRNA Fbxl5 3’-UTR-MUT (Figure 2A and B), in which the AGTTCTCA was replaced with CTGGAGAC. A previous study showed that miRNAs suppress translation and degrade mRNA in an AGO2-dependent manner via binding to their targets.19 Anti-AGO2 immunoprecipitation was conducted in NMVMs overexpressing miR-146a and circRNA Fbxl5, respectively, using anti-AGO2 antibodies or control IgG. It was found that circRNA Fbxl5 was significantly enriched in the biotin-coupled miR-146a-captured fraction, and vice versa (Figure 2C and D). These results demonstrated that circRNA Fbxl5 could function as an efficient miR-146a sponge in cardiomyocytes of mice.
Downregulation of circRNA Fbxl5 Protected Myocardium Against IRI and Cell Death and Preserved Cardiac Function
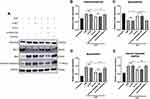
To further indicate whether downregulation of circRNA Fbxl5 has cardioprotective effects in vivo, we adopted an adenoviral-associated vector (AAV9)-troponin T (TnT)-mediated cardiac gene delivery approach, which has been validated in numerous studies. Through tail vein injection of AAV9-TnT-si-circRNA Fbxl5 in the mouse model, we identified significant downregulation of circRNA Fbxl5 in myocardium of mice at the second week after injection. The efficiency of adenovirus transduction was quantified by GFP-positive myocytes, as shown in Supplementary Figure 1. The timeline of the in vivo experiment is shown in Figure 3A. TUNEL staining revealed a significant reduction of apoptosis in IR ventricular myocardium expressing AAV9-TnT-si-circRNA Fbxl5 (Figure 3B). Additionally, Evans blue/TTC staining of ventricular myocardium after IR and echocardiographic analysis showed a smaller infarct size in myocardium and improved cardiac functions expressing AAV9-TnT-si-circRNA Fbxl5 compared with the vehicle and si-NC groups, respectively (Figures 3C-E and J-L). Moreover, Western blotting further validated the increased expression level of Bcl-2 and the decreased expression levels of Bax and cleaved caspase-3 in ventricular myocardium expressing AAV9-TnT-si-circRNA Fbxl5 (Figures 3F-I), which further suggested that knockdown of circRNA Fbxl5 could ameliorate cell death after IR. These results suggest that knockdown of circRNA Fbxl5 attenuated apoptosis and improved cardiac functions induced by IR in vivo.
Downregulation of circRNA Fbxl5 Protected Cardiomyocytes Against AR-Related Cell Death
To explore the effect of circRNA Fbxl5 on cardiomyocyte viability in AR conditions, we manipulated the expression level of circRNA Fbxl5 in NMVMs through siRNA-based knockdown. After transfection for 24 h, the cultured NMVMs were subjected to AR. We found that knockdown of circRNA Fbxl5 attenuated cardiomyocyte apoptosis induced by AR, as evidenced by Western blotting and immunofluorescence, respectively (Figure 4). These results suggested that knockdown of circRNA Fbxl5 attenuated cardiomyocyte apoptosis induced by AR in vitro.
CircRNA Fbxl5 Modulated AR-Related Cell Death Through Regulating the miR-146a-MED1 Axis
Our previous study demonstrated that miR-146a protected against myocardial IRI and attenuated cardiomyocyte apoptosis via targeting MED1.13 In the present study, we identified circRNA Fbxl5, which could function as miR-146a sponge, and aggravate IR-related cardiomyocyte apoptosis. In order to further indicate whether circRNA Fbxl5 could modulate cardiomyocyte apoptosis via regulating the miR-146a-MED1 axis, we manipulated the expression levels of circRNA Fbxl5 and miR-146a in NMVMs through siRNA-based knockdown and miR-146a inhibitor, respectively. As expected, knockdown of circRNA Fbxl5 significantly downregulated the expression level of MED1, and attenuated cardiomyocyte apoptosis induced by AR. Inhibition of miR-146a reversed the effect of knockdown of circRNA Fbxl5 on the expression level of MED1 and cardiomyocyte apoptosis (Figure 5). These results suggested that circRNA Fbxl5 modulated AR-related cell death through regulating the miR-146a-MED1 axis.
Discussion
Noncoding RNAs regulate gene expression post-transcriptionally in various biological processes, as well as cardiovascular diseases. A growing body of evidence demonstrated that miRNAs are involved in gene silencing through degradation of post-transcriptional mRNA or suppression of the protein expression, while circRNAs regulate gene expression mainly through functioning as miRNA sponges.5,20 Studies showed that miR-146a could protect cardiomyocytes against ischemia-induced cell death.21,22 Consistently, our previous research also demonstrated that miR-146a protected against myocardial IRI and attenuated cardiomyocyte apoptosis via targeting MED1.13 However, when we further evaluated the variation of miR-146a in myocardium of mice after IR, we found, for the first time, an interesting phenomenon, in which the expression level of endogenous miR-146a significantly decreased as the duration of IR was prolonged, with the lowest expression level occurred at 1 h after IR. Similarly, an in vitro AR model of NMVMs also recapitulated the results. Thus, we hypothesized that miR-146a could be sponged by upregulating circRNA induced by myocardial IRI.
In order to indicate whether there is a circRNA that can function as miR-146a sponge under myocardial IRI, the following criteria must be met. First, the sequence of this circRNA must have a base-paired region that can target miR-146a; Second, this circRNA must be abundantly expressed in myocardium; Third, myocardial IRI can induce the upregulation of circRNA. To this end, we first predicted top 10 probable circRNAs that can function as miR-146a sponge through TargetScan and miRanda. Then, we established a mouse model of myocardial IRI to evaluate the expression levels of circRNAs in the myocardium. Among them, we identified mmu_circ_0001346, termed as circRNA Fbxl5, which was significantly upregulated at 1 h after IR. Both in vivo and in vitro functional studies demonstrated that circRNA Fbxl5 promoted the apoptosis of cardiomyocytes under IRI. Mechanistically, circRNA Fbxl5 regulates cardiomyocyte apoptosis via targeting miR-146a-MED1 cascade. This is the first study to illustrate that myocardial IRI induces the upregulation of circRNA Fbxl5, acting a sponge for miR-146a to suppress its expression level. The suppression of miR-146a expression level favors the upregulation of MED1, which could further promote cardiomyocyte apoptosis, as shown in our previous study.22
Dysregulated circRNAs contribute to various cardiovascular disorders.23–25 Upregulated expression level of HRCR protected the heart from pathological remodeling and heart failure via sponging miR-223 and increasing ARC expression level.8 CircRNA Ttc3 was significantly induced in the heart after myocardial infarction (MI) and exerted protective effects against cardiomyocyte apoptosis by sponging miR-15b.11 However, MFACR and circRNA ACAP2 promoted cardiomyocyte death during IRI and after MI, respectively.9,23 The present study provided additional insight into the circRNAs mediated during myocardial IRI, which showed for the first time that upregulation of endogenous circRNA Fbxl5 promoted myocardial IRI and cell death by sponging cardioprotective miR-146a. However, further study is required to address the mechanisms of the generation and upregulation of circRNA Fbxl5.
Recently, biocompatible drug delivery vehicles, such as exosomes and nanoparticles, have emerged as a promising therapeutic strategy for alleviating myocardial IRI.26–29 However, due to low yields and complicated purification processes of exosomes, their clinical translation has been hampered.30 Moreover, the onset of MI cannot be predicted clinically. Besides, miR-146a seems to be a promising therapeutic target for ameliorating myocardial IRI, however, the major challenges of RNA therapy are in vivo instability and a relatively low transfection efficiency.31–34 In the present study, we showed that circRNA Fbxl5 was significantly upregulated after IR, which could further sponge the endogenous cardioprotective miR-146a and promote the death of cardiomyocytes. Therefore, we speculate that circRNA sponge may be one of the major pathways, contributing to the instability of both endogenous and exogenous miRNAs. Due to their covalently closed loops, circRNAs exhibit degradation resistance and seems to be a promising biomarker and target for therapeutic intervention. Therefore, targeting circRNA Fbxl5 to prevent the rapid degradation of miR-146a accompanied by an exogenous delivery via biocompatible vehicles seems to be a promising strategy for alleviating myocardial IRI.
The present study has some limitations. First, due to various targets of noncoding RNA, we could not exclude the possibility of the involvement of other circRNAs or other classes of noncoding RNAs in controlling the expression level of miR-146a. Second, the molecular mechanisms responsible for the generation and upregulation of circRNA Fbxl5 need further in-depth studies.
Conclusions
In summary, our study identified for the first time that the expression level of circRNA Fbxl5 significantly increased after myocardial IRI and contributed to the death of cardiomyocytes, thereby aggravating myocardial IRI. Mechanistically, circRNA Fbxl5 sponged the endogenous miR-146a, thereby weakening its inhibitory effect on MED1, which could further promote cardiomyocyte apoptosis. Further study is therefore required to address the mechanisms of the generation and upregulation of circRNA Fbxl5.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (Grant No. 81870264) and the SHIPM-mu fund No. JC202005 from Shanghai Institute of Precision Medicine, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Amani H, Habibey R, Hajmiresmail SJ, Latifi S, Pazoki-Toroudi H, Akhavan O. Antioxidant nanomaterials in advanced diagnoses and treatments of ischemia reperfusion injuries. J Mater Chem B. 2017;5(48):9452–9476. doi:10.1039/C7TB01689A
2. Ibanez B, Heusch G, Ovize M. Van de Werf F. Evolving therapies for myocardial ischemia/reperfusion injury. J Am Coll Cardiol. 2015;65(14):1454–1471. doi:10.1016/j.jacc.2015.02.032
3. Heusch G. Critical Issues for the Translation of Cardioprotection. Circ Res. 2017;120(9):1477–1486. doi:10.1161/CIRCRESAHA.117.310820
4. Hsiao KY, Sun HS, Tsai SJ, Circular RNA. New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141. doi:10.1177/1535370217708978
5. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi:10.1038/nrm.2015.32
6. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
7. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
8. Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi:10.1093/eurheartj/ehv713
9. Wang K, Gan TY, Li N, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24(6):1111–1120. doi:10.1038/cdd.2017.61
10. Zhou LY, Zhai M, Huang Y, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2018;26(7):1299–1315. doi:10.1038/s41418-018-0206-4
11. Cai L, Qi B, Wu X, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. 2019;130:10–22. doi:10.1016/j.yjmcc.2019.03.007
12. Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One. 2016;11(3):e0151753. doi:10.1371/journal.pone.0151753
13. Zhang T, Ma Y, Gao L, et al. MicroRNA-146a protects against myocardial ischaemia reperfusion injury by targeting Med1. Cell Mol Biol Lett. 2019;24:62. doi:10.1186/s11658-019-0186-5
14. National Research Council Committee for the Update of the Guide for the C, Use of Laboratory A. The National Academies Collection: Reports Funded by National Institutes of Health. Guide for the Care and Use of Laboratory Animals. Washington (DC): National Academies Press (US)Copyright © 2011, National Academy of Sciences; 2011.
15. Percie N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 2020;18(7):e3000410. doi:10.1371/journal.pbio.3000410
16. Carroll KJ, Makarewich CA, McAnally J, et al. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113(2):338–343. doi:10.1073/pnas.1523918113
17. Tan WQ, Wang K, Lv DY, Li PF. Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J Biol Chem. 2008;283(44):29730–29739. doi:10.1074/jbc.M805514200
18. Li PF, Li J, Müller EC, Otto A, Dietz R, von Harsdorf R. Phosphorylation by protein kinase CK2: a signaling switch for the caspase-inhibiting protein ARC. Mol Cell. 2002;10(2):247–258. doi:10.1016/S1097-2765(02)00600-7
19. Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi:10.1038/emboj.2011.359
20. Lorenzen J, Kumarswamy R, Dangwal S, Thum T. MicroRNAs in diabetes and diabetes-associated complications. RNA Biol. 2012;9(6):820–827. doi:10.4161/rna.20162
21. Wang X, Ha T, Liu L, et al. Increased expression of microRNA-146a decreases myocardial ischaemia/reperfusion injury. Cardiovasc Res. 2013;97(3):432–442. doi:10.1093/cvr/cvs356
22. Zhang W, Shao M, He X, Wang B, Li Y, Guo X. Overexpression of microRNA-146 protects against oxygen-glucose deprivation/recovery-induced cardiomyocyte apoptosis by inhibiting the NF-κB/TNF-α signaling pathway. Mol Med Rep. 2018;17(1):1913–1918. doi:10.3892/mmr.2017.8073
23. Liu X, Wang M, Li Q, Liu W, Song Q, Jiang H. CircRNA ACAP2 induces myocardial apoptosis after myocardial infarction by sponging miR-29. Minerva Med. 2020;1:45.
24. Si X, Zheng H, Wei G, et al. circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a. Mol Ther Nucleic Acids. 2020;21:636–655. doi:10.1016/j.omtn.2020.06.024
25. Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10(1):4317. doi:10.1038/s41467-019-11777-7
26. Takov K, He Z, Johnston HE, et al. Small extracellular vesicles secreted from human amniotic fluid mesenchymal stromal cells possess cardioprotective and promigratory potential. Basic Res Cardiol. 2020;115(3):26. doi:10.1007/s00395-020-0785-3
27. Geng T, Song ZY, Xing JX, Wang BX, Dai SP, Xu ZS. Exosome Derived from Coronary Serum of Patients with Myocardial Infarction Promotes Angiogenesis Through the miRNA-143/IGF-IR Pathway. Int J Nanomedicine. 2020;15:2647–2658. doi:10.2147/IJN.S242908
28. Sayed N, Tambe P, Kumar P, Jadhav S, Paknikar KM, Gajbhiye V. miRNA transfection via poly(amidoamine)-based delivery vector prevents hypoxia/reperfusion-induced cardiomyocyte apoptosis. Nanomedicine. 2020;15(2):163–181. doi:10.2217/nnm-2019-0363
29. Zhou H, Shan Y, Tong F, et al. Resveratrol Nanoparticle Complex: potential Therapeutic Applications in Myocardial Ischemia Reperfusion Injury. J Biomed Nanotechnol. 2020;16(3):382–389. doi:10.1166/jbn.2020.2900
30. Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:54.
31. Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi:10.1038/nrd2742
32. Zhang Y, Chan HF, Leong KW. Advanced materials and processing for drug delivery: the past and the future. Adv Drug Deliv Rev. 2013;65(1):104–120. doi:10.1016/j.addr.2012.10.003
33. Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi:10.1038/nrg3763
34. Li J, Wu C, Wang W, et al. Structurally modulated codelivery of siRNA and Argonaute 2 for enhanced RNA interference. Proc Natl Acad Sci U S A. 2018;115(12):E2696–e705. doi:10.1073/pnas.1719565115
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.