Back to Journals » OncoTargets and Therapy » Volume 14
Circular RNA circWDR27 Promotes Papillary Thyroid Cancer Progression by Regulating miR-215-5p/TRIM44 Axis
Authors Wang W, Huang C, Luo P, Yao J, Li J, Wang W, Liu F
Received 3 November 2020
Accepted for publication 31 December 2020
Published 19 May 2021 Volume 2021:14 Pages 3281—3293
DOI https://doi.org/10.2147/OTT.S290270
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Weilan Wang,1,* Chengmin Huang,1,* Peng Luo,2 Jiang Yao,1 Jie Li,1 Wenxia Wang,1 Fengqin Liu3
1Department of General Surgery, Changxing People’s Hospital, The Second Affiliated Hospital of Zhejiang University School of Medical Changxing Campus, Changxing, Zhejiang, People’s Republic of China; 2Department of Medical Research, Shanghai Topgen Biomedical Technology Co., Ltd., Shanghai, People’s Republic of China; 3Department of Ultrasound, Changxing People’s Hospital, The Second Affiliated Hospital of Zhejiang University School of Medical Changxing Campus, Changxing, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fengqin Liu
Department of Ultrasound, The Second Affiliated Hospital of Zhejiang University School of Medical Changxing Campus, No. 66 Taihu Middle Road, Changxing, 313100, Zhejiang, People’s Republic of China
Tel +86-0572-6023641
Email [email protected]
Purpose: This study was to explore the biological roles and underlying mechanism of circRNA WD repeat domain 27 (circWDR27).
Methods: The expression of circWDR27, microRNA-215-5p (miR-215-5p) and tripartite motif containing 44 (TRIM44) were measured by quantitative real-time polymerase chain reaction (qRT-PCR). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and colony formation assays were employed to detect cell proliferation. Flow cytometry was used to determine cell apoptosis and cell cycle distribution. Cell migration and invasion abilities were examined by wound healing and transwell assays. The protein levels of matrix metalloproteinase 2 (MMP2), MMP9 and TRIM44 were analyzed by Western blot assay. The relationship between miR-215-5p and circWDR27 or TRIM44 was predicted by bioinformatics tools and confirmed using dual-luciferase reporter assay. Mouse xenograft model was established to examine the role of circWDR27 in vivo.
Results: CircWDR27 and TRIM44 were highly expressed while miR-215-5p was lowly expressed in PTC tissues and cells. Knockdown of circWDR27 suppressed cell proliferation and metastasis and induced cell cycle arrest and apoptosis in PTC cells. Moreover, miR-215-5p was a direct target of circWDR27, and its inhibition reversed the suppressive effect of circWDR27 knockdown on PTC cell progression. In addition, miR-215-5p directly targeted TRIM44, and miR-215-5p exerted its anti-cancer role in PTC cells by targeting TRIM44. Furthermore, circWDR27 positively regulated TRIM44 expression by sponging miR-215-5p. Importantly, knockdown of circWDR27 suppressed tumor growth in vivo by upregulating miR-215-5p and downregulating TRIM44.
Conclusion: CircWDR27 accelerates PTC progression via regulating miR-215-5p/TRIM44 axis, providing a potential therapeutic target for PTC.
Keywords: papillary thyroid cancer, circWDR27, miR-215– 5p, TRIM44
Introduction
Thyroid cancer (TC) is the most common endocrine malignant tumor worldwide, and its incidence has steadily increased in the past few decades.1 The global incidence rate of TC in women of 10.2 per 100,000 is 3 times higher than in men.2 Papillary thyroid cancer (PTC) accounts for 80–90% of all TC.3 Most patients with PTC have a generally good prognosis with the current therapeutic regimen, including surgical resection, radioactive iodine therapy, and thyroid hormone suppression.4 Nevertheless, a small proportion of patients with PTC have poor prognosis due to large primary tumor, lymph node metastasis, and distant metastasis.5 Hence, it is imperative to explore the pathogenesis of PTC, and thus developing effective therapeutic targets to improve the clinical outcomes.
Circular RNAs (circRNAs), a novel type of noncoding RNA (ncRNA), have special closed-loop structures without 5ʹ-cap and 3ʹ–end poly A tail, which make them more stable and resistant to RNase R.6 Increasing studies have demonstrated that circRNAs are widely involved in diverse physiological and pathological processes.7 Moreover, dysregulation of circRNAs is correlated with the progression of many cancers, including PTC.8 For instance, circ_0067934 upregulation was correlated with poor prognosis and promoted PTC progression.9 Moreover, hsa_circ_0058124 accelerated PTC tumorigenesis by regulating NOTCH3/GATAD2A axis.10 CircRNA WD repeat domain 27 (circWDR27, also known as hsa_circ_0078738) is derived from of WDR27 gene and located at chr6:170033042-170058454, and circWDR27 is upregulated in PTC tissues.11 However, the exact roles and regulatory mechanism of circWDR27 in PTC remain largely unknown.
Many circRNAs have been demonstrated to contain several binding sites of microRNA (miRNA). Therefore, they can be used as RNA “sponges” to adsorb miRNAs, thereby protecting target genes from miRNA-mediated mRNA degradation through the mechanisms of competition for endogenous RNA (ceRNAs).12,13 In tumor research, circRNAs participate in tumor progression by sponging miRNAs to affect the expression of target genes.14,15 Recent researches revealed that miR-215–5p acted as a tumor suppressor in many cancers, including PTC.16–18 Moreover, tripartite motif containing 44 (TRIM44) has been reported to play a carcinogenic role in PTC.19 Interestingly, online bioinformatics database showed that circWDR27 and TRIM44 had complementary binding sequence for miR-215-5p. Based on these findings, we assumed the ceRNA network of circWDR27/miR-215-5p/TRIM44 in PTC.
In this research, we analyzed the expression of circWDR27 in PTC tissues and cells. Functionally, we measured the impact of circWDR27 on cell growth, apoptosis, cell cycle, and metastasis. Mechanistically, we predicted and validated that the miR-215-5p/TRIM44 axis was the downstream target of circWDR27. The aim of this study is to provide novel evidence for the development of clinical therapeutic strategies against PTC.
Materials and Methods
Specimen Collection
A total of 42 pairs of PTC tissues and adjacent normal tissues were obtained from patients who had undergone surgery at Changxing people’s hospital between January 2018 to June 2019. All samples were confirmed by pathological examination in our hospital. None of the patients with PTC received radiotherapy or any other treatment before the operation. All tissue specimens were timely snap-frozen in liquid nitrogen and then kept in a refrigerator at −80°C until RNA or protein extraction. Informed consents were obtained from all patients. This research was permitted by the Research Ethics Committee of Changxing people’s hospital, which was in accordance with the Declaration of Helsinki Principles.
Cell Culture and Transfection
Human thyroid follicular epithelial cells (Nthy-ori3-1) and human PTC cells (TPC-1 and IHH-4) were acquired from COBIOER (Nanjing, China). These cells were cultured in RPMI-1640 medium (Cat. No.SH30027.01, HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Cat. No.10099141, Invitrogen, Carlsbad, CA, USA) at 37°C in an atmosphere of 5% CO2.
Small interfering RNA (siRNA) against circWDR27 (si-circWDR27), miR-215-5p mimics (miR-215-5p), miR-215-5p inhibitor (anti-miR-215-5p), TRIM44 overexpression vector (TRIM44), and their matched controls (si-NC, miR-NC, anti-miR-NC, vector) were bought from Genecreat (Wuhan, China). Lentivirus-mediated short hairpin RNA (shRNA) targeting circWDR27 (sh-circWDR27) and corresponding control (sh-NC) were obtained from RiboBio (Guangzhou, China). The full-length sequence of TRIM44 was cloned into the pcDNA3.1 vector to obtain the overexpression vector for TRIM44. The sequences were as follows: si-circWDR27 (5ʹ-AAAATAAACTCCTGTGTCCTA-3ʹ); si-NC (5ʹ-GCGCGATAGCGCGAATATA-3ʹ); miR-215-5p mimics (5ʹ-AAUGACCUAUGAAUUGACAGAC-3ʹ); miR-NC (5ʹ-ACUCUAUCUGCACGCUGACUU-3ʹ); miR-215-5p inhibitor (5ʹ-GUCUGUCAAUUCAUAGGUCAU-3ʹ); anti-miR-NC (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ), sh-circWDR27 (5ʹ-CCGGGCCAAAATAAACTCCTGTGTCCTCGAGGACACAGGAGTTTATTTTGGCTTTTTG-3ʹ); (5ʹ-CCGGAACGAGTTCAACTCGTTACCCTCGAGGTAACGAGTTGAACTCGTTTTTTTG-3ʹ). Cell transfection was performed by using Lipofectamine 3000 (Cat.No.L3000015, Invitrogen).
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Tissue samples or cells were subjected to TRIzol reagent (Cat. No, 5596‐018, Invitrogen) to isolate total RNA. The complementary DNA (cDNA) was synthesized by PrimeScriptTM RT reagent Kit (Cat.No.RR037A, TaKaRa, Dalian, China) or miRNA cDNA synthesis kit (Cat.No.D350A, TaKaRa). Next, qRT-PCR was conducted with SYBR Green PCR Master Mix Kit (Cat.No.RR820W, TaKaRa) under the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The expression of genes was analyzed with the 2−ΔΔCt method, followed by normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; for circWDR27, WDR27 and TRIM44) or U6 (for miR-215-5p). The following primers were used: circWDR27 (sense, 5ʹ-GAACAGGACCGTGGAAGTGT-3ʹ; anti-sense, 5ʹ-GAGGTCGGTAGGACACAGGA-3ʹ); WDR27 (sense, 5ʹ-CTCCACTGTATCTGGGAATTGCC-3ʹ; anti-sense, 5ʹ-AACCAGGCGTTGGTCCTTCATG-3ʹ); miR-215-5p (sense, 5ʹ-GCCGAGATGACCTATGAAT-3ʹ; anti-sense, 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ); TRIM44 (sense, 5ʹ-GTGGACATCCAAGAGGCAAT-3ʹ; anti-sense, 5ʹ-AGCAAGCCTTCATGTGTCCT-3ʹ); GAPDH (sense, 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ; anti-sense, 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ), U6 (sense, 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ; anti-sense, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
RNase R Treatment
RNase R (Cat.No.RNR07250, Epicentre Technologies, Madison, WI, USA) treatment was used to degrade linear RNA. In brief, total RNA (2 μg) was incubated with or without RNase R (3 U/μg) for 0.5 h at 37°C. After that, the treated RNA was used for qRT-PCR to examine the levels of circWDR27 and WDR27.
Subcellular Fractionation Location
Referring to manufacturer’s instructions, PARIS™ Kit (Cat.No.AM1921, Invitrogen) was used to isolate the RNA from cytosolic and nuclear fractions. The extracted RNAs from nuclear and cytosolic fractions were analyzed using the qRT-PCR analysis. U6 or GAPDH was functioned as a nucleus control or cytoplasm control, respectively.
Cell Proliferation Assays
Cell viability was estimated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, TPC-1 and IHH-4 cells were cultured in a 96–well plate. At different points after transfection, MTT reagent (Cat.No.ST316, Beyotime, Jiangsu, China) was added into per well and incubated for 3-4 h. After removing the medium, dimethyl sulfoxide (DMSO; Cat.No.ST038 150 μL, Beyotime) solution was added to each well. Finally, the absorbance at 570 nm wavelength was measured using a microplate reader (Bio-Rad).
For clone formation assay, transfected cells (PC-1 and IHH-4) were inoculated in a 6-well plate and then cultured for 2 weeks. Afterwards, cells were fixed by paraformaldehyde (Cat.No.P0099, Beyotime) for 0.5 h, and stained by crystal violet (Cat.No.C0121, Beyotime) for 2 h. Finally, the number of colonies (>50 cells per colony) was counted by microscope (Leica, Wetzlar, Germany).
Flow Cytometry Analysis
For apoptosis analysis, Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Cat.No.KGA107, Keygen, Nanjing, China) was administrated for the apoptosis detection. In brief, cells were collected and washed, and then incubated with Annexin V-FITC (10µL) and PI (5 µL). At last, flow cytometry (BD Biosciences, Franklin, NJ, USA) was used for measuring cell apoptosis.
For cell cycle determination, TPC-1 and IHH-4 cells were collected and fixed with ice-cold 70% ethanol (Cat.No.A500737, Sangon Biotech, Shanghai, China) at −20°C for 12 h. Next, the cells were collected and washed, followed by incubation with PI (Cat.No.A601112, 25 μg/mL, Sangon Biotech) and 100 µg/mL RNase A (Cat.No.B600476, Sangon Biotech) for 15 min. At last, flow cytometer was applied to detect cell cycle distribution.
Wound Healing and Transwell Assays
Wound healing assay was performed to detect cell migration ability. In short, transfected cells (PC-1 and IHH-4) were cultured in 12-well plates. At ~80% confluence, cells were gently scratched with a sterile pipette tip (20 μL). Images were captured using a microscope (Leica, ×40) at 0 and 24 h after scratching. The distance of cell migration was measured using ImageJ software.
For transwell assay, transfected cells (PC-1 and IHH-4) in 0.2 mL serum-free medium were added to the top chamber (Costar, Corning, NY, USA) with (invasion) or without (migration) Matrigel (BD Biosciences), and 0.6 mL DMEM medium with 10% FBS was added to the bottom chamber. The non-migrating/invading cells from the top surface of the insert were gently removed after 24 h incubation. And migrating/invading cells from the bottom side of the membrane were fixed by paraformaldehyde (Beyotime) for 0.5 h, and stained by crystal violet (Beyotime) for 2 h. Then, the stained cells were counted and photographed with a microscope (Leica) at magnification of ×100.
Western Blot Assay
The proteins were extracted using RIPA lysis buffer (Cat.No.KGP702, Keygen) with 1% protease inhibitor phenylmethanesulfonyl fluoride (Cat.No.ST505, Beyotime). The proteins were added to 2 × loading buffer (Cat.No.P0015B, Beyotime) and denatured by heating at 100°C for 5 min, followed by quantification with BCA protein assay kit (Cat.No.ab102536, Abcam, Cambridge, UK). Total proteins (about 40 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Cat.No.P0670, Beyotime), and then electrophoretically transferred to 0.2 μM polyvinylidene fluoride (PVDF) membranes (Cat.No.FFP24, Beyotime). Then, the membranes were blocked in 5% non–fat milk (Cat.No.A600669, Sangon Biotech) for 1.5–2 h, followed by incubation with the appropriate primary antibody at 4°Cfor 12–16 h. Afterwards, all membranes were maintained in secondary antibody (Cat.No.D110058, 1:4000, Sangon Biotech). Finally, the protein signals were analyzed using enhanced chemiluminescence kit (Cat.No.P1000, Applygen, Beijing, China). The primary antibodies were purchased from Abcam and listed as below: matrix metalloproteinase 2 (MMP2; Cat.No.ab235167, 1:1000), MMP9 (Cat.No.ab38898, 1:1000), TRIM44 (Cat.No.ab236422, 1:2000), β-actin (Cat.No.ab227387, 1:5000).
Dual-Luciferase Reporter Assay
The putative binding sites of miR-215-5p and circWDR27 or TRIM44 were predicted by circinteractome or starBase v2.0. To construct wild-type (WT) luciferase reporter vector (circWDR27 WT or TRIM44 3ʹUTR WT), the fragments of circWDR27 or TRIM44 3ʹUTR containing the predicted binding sites for miR-215-5p were synthesized and inserted into XhoI and NotI sites of the pmiR-RB-REPORT™ luciferase reporter vector (Cat.NoPMIR1001, RiboBio). To construct mutant (WT) luciferase reporter vector (circWDR27 MUT or TRIM44 3ʹUTR MUT), circWDR27 mutant sequence or TRIM44 3ʹUTR mutant sequence, in which several nucleotides were mutated within the miR-215-5p binding sites, was also inserted into the vector. MiR-215-5p or miR-NC and above reporter plasmid (WT or MUT) were co-transfected into TPC-1 and IHH-4 cells for 48 h. Next, dual–luciferase reporter assay system (Promega, Madison, WI, USA) was used for measuring the luciferase activity.
Tumor Formation Assay in vivo
BALB/c nude mice (female, n=12, 5–6 weeks old, weighing 18-25 g, Vital River Laboratory Animal, Inc., Beijing, China) were used for the in vivo tumor formation assay. TPC-1 cells (2 × 106) transiently transfected with sh-NC (as control) or sh-circWDR27 were injected into the right flank of nude mice (n=6/group). Tumor volume (V) was monitored and recorded via examining the width (W) and length (L) of tumors every 7 days and calculated according to the equation: V = L× W2/2. After 35 days of inoculation, the mice were sacrificed, and subcutaneous tumors from sacrificed mice were removed for further testing. The in vivo experiments were approved by the Animal Care and Use Committee of Changxing people’s hospital and performed according to the guidelines for laboratory animal welfare (GB/T 35,892–2018).
Statistical Analysis
Graph Prism 6.0 software (GraphPad Prism, San Diego, CA, USA) was used for data analysis. Data were expressed as the mean ± standard deviation (SD) from at least three independent experiments. Statistical significance was analyzed via Student’s t-test (for two groups) or a one-way analysis of variance (ANOVA; for multiple groups). Pearson’s correlation coefficient was employed to confirm the correlations among circWDR27, miR-215-5p, and TRIM44. For all comparisons, statistical significance was considered when P<0.05.
Results
CircWDR27 Was Upregulated in PTC Tissues and Cells
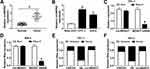
To explore whether the expression of circWDR27 was dysregulated in PTC, qRT-PCR was performed. As displayed in Figure 1A, circWDR27 expression was obviously higher in PTC tissues than that in the corresponding adjacent normal tissues. Similarly, circWDR27 level was enhanced in PTC cells (TPC-1 and IHH-4) relative to Nthy-ori3-1 cells (Figure 1B). Next, the characteristics of circWDR27 in PTC cells were analyzed. The data from qRT-PCR showed that circWDR27 was resistant to RNase R digestion (Figure 1C and D), suggesting the cyclic structure of circWDR27. Subsequently, the localization of circWDR27 was examined in TPC-1 and IHH-4 cells. As presented in Figure 1E and F, circWDR27 was predominantly located in the cytoplasm. These results indicated that circWDR27 was upregulated in PTC and had a loop structure.
Knockdown of circWDR27 Inhibited Cell Proliferation and Metastasis and Induced Apoptosis and Cell Cycle Arrest in PTC Cells
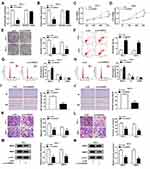
In order to figure out the precise role of circWDR27 in PTC, TPC-1 and IHH-4 cells were transfected with si-NC or si-circWDR27. The expression of circWDR27 was reduced in both TPC-1 and IHH-4 cells under the transfection of si-circWDR27 in comparison to that in those cells transfected with si-NC, but the mRNA expression of WDR27 was not affected by transfection of si-circWDR27 (Figure 2A and B). MTT assay showed that cell viability was reduced after knockdown of circWDR27 in TPC-1 and IHH-4 cells (Figure 2C and D). Colony formation assay demonstrated that downregulation of circWDR27 decreased the number of colonies (Figure 2E). Flow cytometry analysis showed silence of circWDR27 induced apoptosis of TPC-1 and IHH-4 cells, and increased the proportions of TPC-1 and IHH-4 cells in G0/G1 phase, while decreased the proportions of these cells in S phase, indicating that the cell cycle was arrested at the G0/G1 phase (Figure 2F–H). Wound healing assay showed that inhibition of circWDR27 reduced wound healing rate in TPC-1 and IHH-4 cells, suggesting that circWDR27 knockdown inhibited TPC-1 and IHH-4 cell migration (Figure 2I and J). Meanwhile, transwell assay showed that interference of circWDR27 suppressed TPC-1 and IHH-4 cell migration and invasion (Figure 2K and L). MMP2 and MMP9 are believed to play important roles in PTC metastasis.20 Western blot assay indicated that knockdown of circWDR27 reduced the protein levels of MMP 2 and MMP 9 in TPC-1 and IHH-4 cells (Figure 2M and N). Collectively, these data indicated that circWDR27 played a growth-promoting role in PTC development.
CircWDR27 Acted as a Sponge of miR-215-5p
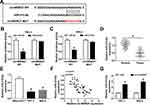
It is well known that circRNAs can exert their functions through binding with their downstream miRNAs. Thus, we searched for the predicted target miRNAs of circWDR27 using the circinteractome. As showed in Figure 3A, circWDR27 contained potential binding sites of miR-215-5p. Next, dual-luciferase reporter assay was conducted to verify the targeting relationship between circWDR27 and miR-215-5p. Luciferase reporter assay results showed that TPC-1 and IHH-4 cells co-transfected with miR-215-5p and circWDR27 WT had lower luciferase activity than that of cells co-transfected with miR-NC and circWDR27 WT, however, no evident change on the luciferase activity was observed in cells transfected with a mutant circWDR27 binding sequence (Figure 3B and C). Subsequently, we measured the expression of miR-215-5p in PTC tissues and cells. As presented in Figure 3D and E, the expression of miR-215-5p was reduced in PTC tissues and cells compared to normal tissues and cells. Moreover, we observed that the expression of circWDR27 was negatively correlated with the level of miR-215-5p in PTC tissues (Figure 3F). Then, we investigated the impact of circWDR27 on miR-215-5p expression. As shown in Figure 3G, knockdown of circWDR27 promoted the expression of miR-215-5p in TPC-1 and IHH-4 cells. All these data indicated miR-215-5p was a target of circWDR27.
Downregulation of circWDR27 Inhibited PTC Cell Progression by Sponging miR-215-5p
Based on the above findings, we speculated that circWDR27 knockdown inhibited PTC cell progression via regulating miR-215-5p. To validate this hypothesis, TPC-1 and IHH-4 cells were transfected with si-NC, si-circWDR27, si-circWDR27 + anti-miR-NC, or si-circWDR27 + anti-miR-215-5p. The expression of miR-215-5p was increased by knockdown of circWDR27, which was reversed by downregulating miR-215-5p (Figure 4A). Moreover, the inhibitory effects of circWDR27 silence on cell viability and colony formation were abolished by inhibition of miR-215-5p (Figure 4B–D). In addition, silence of miR-215-5p weakened si-circWDR27-induced apoptosis in TPC-1 and IHH-4 cells (Figure 4E). Furthermore, knockdown of miR-215-5p could reverse circWDR27 silence-mediated promotion of G0/G1 phase cells and reduction of S phase cells (Figure 4F and G). Besides, miR-215-5p interference abated the anti-migration and anti-invasion effects caused by knockdown of circWDR27 in TPC-1 and IHH-4 cells (Figure 4H–J). Meanwhile, the suppressive effect of si-circWDR27 on MMP2 and MMP9 protein expression was abated by downregulation of miR-215-5p (Figure 4K and L). Taken together, these findings implied that circWDR27 exerted its tumor oncogenic roles through regulating miR-215-5p.
TRIM44 Was a Direct Target of miR-215-5p
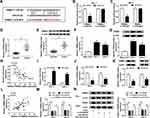
To further elucidate the mechanism of miR-215-5p, target prediction was performed. By searching online software (starBase v2.0), TRIM44 was predicted as a potential target gene of miR-215-5p (Figure 5A). Next, the targeting relationship between TRIM44 and miR-215-5p was identified using the dual-luciferase reporter assay. The result revealed that overexpression of miR-215-5p suppressed the luciferase activity of TRIM44 3ʹUTR WT, whereas the luciferase activity of TRIM44 3ʹUTR MUT was not evidently affected by upregulating miR-215-5p (Figure 5B and C). Moreover, the expression of TRIM44 in PTC tissues and cells was examined. We found that the mRNA and protein expression of TRIM44 were both increased in PTC tissues relative to normal tissues (Figure 5D and E). Likewise, high mRNA and protein expression of TRIM44 were observed in PTC cells (TPC-1 and IHH-4) compared to Nthy-ori3-1 cells (Figure 5F and G). And we found that miR-215-5p expression was negatively correlated with TRIM44 mRNA level in PTC tissues (Figure 5H). Overexpression efficiency of miR-215-5p was validated by qRT-PCR (Figure 5I). Subsequently, the influence of miR-215-5p on the expression of TRIM44 was explored. As illustrated in Figure 5J and K, overexpression of miR-215-5p inhibited TRIM44 mRNA and protein levels in TPC-1 and IHH-4 cells. Furthermore, circWDR27 expression was positively correlated with TRIM44 mRNA expression in PTC tissues (Figure 5L). Next, we explored whether circWDR27 acted as a sponge of miR-215-5p to regulate TRIM44 in PTC cells. Results showed that circWDR27 interference reduced the mRNA and protein expression of TRIM44 in TPC-1 and IHH-4 cells, which was restored by knockdown of miR-215-5p (Figure 5M and N), indicating that circWDR27 positively regulated TRIM44 expression via sponging miR-215-5p.
Overexpression of miR-215-5p Suppressed PTC Cell Progression by Targeting TRIM44
To investigate whether TRIM44 was involved in miR-215-5p-mediated functions in PTC cells, TPC-1 and IHH-4 cells were transfected with miR-NC, miR-215-5p, miR-215-5p + vector, or miR-215-5p +TRIM44. The protein expression of TRIM44 was reduced by overexpressing miR-215-5p, which was rescued by upregulating TRIM44 (Figure 6A). Moreover, overexpression of miR-215-5p decreased cell viability and colony formation and induced cell apoptosis and cell cycle arrest at G0/G1 phase, while these effects were reversed by elevation of TRIM44 (Figure 6B–G). In addition, enforced expression of miR-215-5p suppressed cell migration and invasion, which was reversed by upregulating TRIM44 (Figure 6H–J). Western blot assay showed that MMP2 and MMP9 protein levels were decreased by transfection with miR-215-5p, which could be abolished by overexpression of TRIM44 (Figure 6K and L). Taken together, these findings demonstrated that miR-215-5p exerted its anti-cancer role in PTC cells via downregulating TRIM44.
Interference of circWDR27 Repressed Tumor Growth in vivo
Next, we explored the effect of circWDR27 on PTC in vivo, sh-NC or sh-circWDR27-transfected TPC-1 cells were injected into the flank of nude mice. Results showed that tumor volume and weight in the sh-circWDR27 group were smaller than those in sh-NC group (Figure 7A and B). The images of tumor tissues were presented in Figure 7C. Moreover, circWDR27 inhibition decreased the expression of circWDR27 and increased the expression of miR-215-5p in excised tumor tissues (Figure 7D and E). Furthermore, silencing circWDR27 reduced the protein level of TRIM44 in tumor tissues (Figure 7F). Altogether, these data proved that circWDR27 knockdown limited tumor growth via upregulating miR-215-5p and downregulating TRIM44.
Discussion
Understanding the critical functions and underlying mechanisms of circRNAs is becoming a novel focus in cancer research.21 However, few circRNAs have been well characterized in terms of function and mechanism in PTC. Herein, we found that circWDR27 accelerated cell proliferation and metastasis and induced apoptosis and cell cycle arrest in PTC. Mechanistically, circWDR27 served as as a ceRNA for miR-215-5p to upregulate TRIM44 expression in PTC.
A large number of studies have shown that circRNAs are considered as promising biomarkers for the diagnosis and prognosis of a variety of cancers due to their abundance and stability in plasma and tissues.22 Previous studies have shown that several circRNAs participate in regulating cell phenotypes of PTC cells through different pathways. For example, inhibition of circBACH decreased PTC cell growth and metastasis through sponging miR-139-5p and downregulating LMO4.23 Moreover, circ-ITCH upregulation suppresses the proliferation and invasion and accelerated cell apoptosis in PTC cells via modulating miR-22-3p/CBL/β-catenin pathway.24 Although circWDR27 has been reported to be highly expressed in PTC tissues, the biological role of circWDR27 in PTC has not been elucidated.11 In this research, we also observed that circWDR27 level was elevated in PTC tissues and cells. Further functional experiments showed that circWDR27 downregulation inhibited the progression of PTC. These findings revealed that circWDR27 acted as an oncogene in PTC.
CircRNAs are well known to function as miRNA sponges and cause the loss of miRNA functions, thereby further promoting the expression of their target genes.25 To explore whether circWDR27 regulate PTC progression via sponging miRNAs, bioinformatics analysis was used. The results demonstrated that miR-215-5p was a direct target of circWDR27. MiR-215-5p has been suggested to act as a tumor-suppressive miRNA in multiple tumors. For example, miR-215-5p upregulation limited growth and migration of colorectal cancer cells.26 Moreover, miR-215-5p inhibited prostate cancer metastasis through regulating PGK1.27 More importantly, miR-215 was reported to be downregulated in PTC cells and tissues, and enforced expression of miR-215 repressed PTC cell proliferation and metastasis by targeting ARFGEF1 and regulating AKT/GSK-3β/Snail signaling.18 Our study revealed that miR-215-5p silence abated the influence of si-circWDR27 on cell proliferation, apoptosis, cell cycle, and metastasis. Consequently, circWDR27 promoted PTC cell progression by decreasing miR-215-5p expression.
The downstream targets of miR-215-5p were also detected. Our results showed that miR-215-5p could directly bind with TRIM44. Previous studies have revealed that TRIM44 plays key roles in various physiological processes, including cell growth, migration, autophagy, apoptosis, and innate immunity.28,29 Moreover, TRIM44 acts as a tumor promoter in diverse cancers, including lung cancer,30 esophageal cancer,31 and gastric carcinoma.28 Additionally, TRIM44 has been shown to be overexpressed in PTC tissues and cells, and TRIM44 inhibition can restrain the proliferation and invasion in PTC cells via suppressing the Wnt/β-catenin signaling pathway.19 In line with this research, TRIM44 overexpression could abolish the anti-cancer role of miR-215-5p in PTC cells, implying the tumorigenic role of TRIM44.
Conclusion
In conclusion, knockdown of circWDR27 could inhibit PTC progression via modulating miR-215-5p/TRIM44 axis, indicating that targeting the circWDR27/miR-215-5p/TRIM44 axis might be a new therapeutic strategy for PTC. Moreover, our study is the first to identify the role of circWDR27, which may provide an effective biomarker for diagnosis and prognosis of PTC.
Highlights
- CircWDR27 is upregulated in PTC tissues and cells.
- CircWDR27 silence suppresses the progression of PTC by increasing miR-215-5p expression.
- MiR-215-5p exerts the anti-cancer role in PTC cells by decreasing TRIM44 expression.
- CircWDR27 acts as a miR-215-5p sponge to regulate TRIM44 expression.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Changxing People’s Hospital. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Acknowledgment
Weilan Wang and Chengmin Huang are co-first authors.
Funding
No funding was received.
Disclosure
The authors declare that they have no competing interests.
References
1. Kitahara CM, Sosa JA. The changing incidence of thyroid cancer. Nat Rev Endocrinol. 2016;12(11):646–653. doi:10.1038/nrendo.2016.110
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Carling T, Udelsman R. Thyroid cancer. Annu Rev Med. 2014;65:125–137. doi:10.1146/annurev-med-061512-105739
4. Fröhlich E, Wahl R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat Rev. 2014;40(5):665–674. doi:10.1016/j.ctrv.2014.01.002
5. Blomberg M, Feldt-Rasmussen U, Andersen KK, Kjaer SK. Thyroid cancer in Denmark 1943–2008, before and after iodine supplementation. Int J Cancer. 2012;131(10):2360–2366. doi:10.1002/ijc.27497
6. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
7. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi:10.1080/15476286.2016.1220473
8. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
9. Wang H, Yan X, Zhang H, Zhan X. CircRNA circ_0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med Sci Monit. 2019;25:1342–1349. doi:10.12659/msm.913463
10. Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 2019;38(1):318. doi:10.1186/s13046-019-1321-x
11. Ye M, Hou H, Shen M, Dong S, Zhang T. Circular RNA circFOXM1 plays a role in papillary thyroid carcinoma by sponging miR-1179 and regulating HMGB1 expression. Mol Ther Nucleic Acids. 2020;19:741–750. doi:10.1016/j.omtn.2019.12.014
12. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
13. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
14. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. doi:10.1038/s41419-019-1382-y
15. Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi:10.1038/s41419-019-2089-9
16. Gao JB, Zhu MN, Zhu XL. miRNA-215-5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio. 2019;9(11):1957–1967. doi:10.1002/2211-5463.12733
17. Yan J, Wei R, Li H, Dou Y, Wang J. miR-452-5p and miR-215-5p expression levels in colorectal cancer tissues and their relationship with clinicopathological features. Oncol Lett. 2020;20(3):2955–2961. doi:10.3892/ol.2020.11845
18. Han J, Zhang M, Nie C, et al. miR-215 suppresses papillary thyroid cancer proliferation, migration, and invasion through the AKT/GSK-3β/Snail signaling by targeting ARFGEF1. Cell Death Dis. 2019;10(3):195. doi:10.1038/s41419-019-1444-1
19. Zhou Z, Liu Y, Ma M, Chang L. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:98–103. doi:10.1016/j.biopha.2017.09.132
20. Wu W, Zhou Q, Zhao W, et al. Ginsenoside Rg3 inhibition of thyroid cancer metastasis is associated with alternation of actin skeleton. J Med Food. 2018;21(9):849–857. doi:10.1089/jmf.2017.4144
21. Zhu LP, He YJ, Hou JC, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37(5). doi:10.1042/bsr20170750
22. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
23. Cai X, Zhao Z, Dong J, et al. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10(3):184. doi:10.1038/s41419-019-1439-y
24. Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem Biophys Res Commun. 2018;504(1):283–288. doi:10.1016/j.bbrc.2018.08.175
25. Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi:10.3389/fmolb.2017.00038
26. Vychytilova-Faltejskova P, Merhautova J, Machackova T, et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 2017;6(11):399. doi:10.1038/s41389-017-0006-6
27. Chen JY, Xu LF, Hu HL, Wen YQ, Chen D, Liu WH. MiRNA-215-5p alleviates the metastasis of prostate cancer by targeting PGK1. Eur Rev Med Pharmacol Sci. 2020;24(2):639–646. doi:10.26355/eurrev_202001_20040
28. Kashimoto K, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012;103(11):2021–2026. doi:10.1111/j.1349-7006.2012.02407.x
29. Yamada Y, Takayama KI, Fujimura T, et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci. 2017;108(1):32–41. doi:10.1111/cas.13105
30. Luo Q, Lin H, Ye X, Huang J, Lu S, Xu L. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF-κB signaling pathway. Int J Clin Oncol. 2015;20(3):508–517. doi:10.1007/s10147-014-0752-9
31. Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018;109(10):3080–3092. doi:10.1111/cas.13762
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.