Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA circTUBGCP3 Is Up-Regulated and Promotes Cell Proliferation, Migration and Survivability via Sponge mir-30b in Osteosarcoma
Authors Xu Y, Yao T, Huang K, Liu G, Huang Y, Gao J, Ye H, Shen S, Ma J
Received 9 January 2020
Accepted for publication 19 April 2020
Published 4 May 2020 Volume 2020:13 Pages 3729—3737
DOI https://doi.org/10.2147/OTT.S245366
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Yining Xu,1,2,* Teng Yao,2,* Kangmao Huang,2,* Gang Liu,2 Yizhen Huang,2 Jun Gao,2 Huali Ye,2 Shuying Shen,2 Jianjun Ma1,2
1Shaoxing University School of Medicine, Shaoxing, Zhejiang 312000, People’s Republic of China; 2Department of Orthopaedic Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine & Key Laboratory of Musculoskeletal System Degeneration and Regeneration Translational Research of Zhejiang Province, Hangzhou, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shuying Shen
Department of Orthopaedic Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine & Key Laboratory of Musculoskeletal System Degeneration and Regeneration Translational Research of Zhejiang Province, Hangzhou, Zhejiang 310000, People’s Republic of China
Tel +0086-057186002200
Fax +0086-057186044817
Email [email protected]
Jianjun Ma
Shaoxing University School of Medicine, Shaoxing, Zhejiang 312000, People’s Republic of China
Email [email protected]
Purpose: Prevailing evidences have demonstrated that circular RNAs (circRNAs) are closely associated with various stages of carcinogenesis. However, very few studies have delineated the specific mechanism of association between circRNAs and osteosarcoma (OS). It offers a novel insight that circRNAs can be explored as a potential therapeutic strategy for OS.
Materials and Methods: In this study, circTUBGCP3 was chosen from the existing reported circRNA microarray data obtained from OS cell lines and normal bone cells. Subsequently, qRT-PCR was performed to evaluate the expression level of circTUBGCP3 in OS samples and cell lines. Functional assays were conducted to estimate the impact of circTUBGCP3 on human OS cells proliferation, vitality, survivability, and migration. Western blot, luciferase reporter and in vivo tumorigenesis assays were performed to analyze the signaling pathways underlying the interaction of circTUBGCP3, miR-30b, and Vimentin.
Results: The data indicate that circTUBGCP3 may act as a sponge of miR-30b that further alters the expression of Vimentin, and promotes the proliferation and metastatic properties of OS cells.
Conclusion: circTUBGCP3 serves as a tumor promoter in tumorigenesis by increasing the possibilities of OS initiation and proliferation.
Keywords: osteosarcoma, circular RNA, circTUBGCP3, miR-30b, Vimentin
Introduction
Osteosarcoma (OS) is a malignant cancer of bones primarily diagnosed during childhood and adolescence with a severe prognosis.1–3 The incidence rate of Osteosarcoma has increased drastically worldwide over the past few decades. The main effective therapy for OS is tumor excision combined with chemo- and radiotherapy.4,5 Unfortunately, because of the high metastasis and invasiveness of OS, the prognosis of osteosarcoma patients with advanced stages is unfavorable.5 Genetically, Osteosarcoma is considered a flawed differentiation disease that is caused by genetic or epigenetic damages during osteogenic differentiation.6 Therefore, it is essential to improve our understanding of osteosarcoma biology and its molecular pathogenesis for better diagnosis and prognosis.
Circular RNAs (circRNAs) are non-coding RNAs with covalent closed loop structure without 5’ end cap and 3’ end poly (A) tail.7 Absence of sensitivity to exonuclease and ribonuclease results in structural conservation and sequence stability of circRNAs.8 Consequently, circRNAs may serve as an ideal tumor biomarker and potential therapeutic targets.
CircRNAs have been widely studied, as they are closely associated with the occurrence and development of cancers. Recently, for the first time Guan et al,9 through microarray analysis, found that hsa_circ_0016788 is highly expressed in liver cancer tissues and accelerates the proliferation of hepatocellular carcinoma cells. Previous studies have discovered the presence of several OS-related circRNAs such as circNASP, circHIPK3, circNT5C2, and circANKIB1.10,13 For instance, one study has shown that the expression of circHIPK3 is down regulated in OS cell lines, tissues, and plasma.13 Thus, we strongly believe that circRNAs have great potential to be explored as novel targets for the treatment of osteosarcoma.
Vimentin, an abundant and highly conserved epithelial-mesenchymal transition protein, is a major member of the type III intermediate filament (IF) protein family.14 It is considered to maintain the integrity and the motility of cells during cell migration and invasion.15 A recent study has shown that Vimentin participates in various complex biological functions in different states of physiology and pathology.16 Further, it is closely related to the invasive and metastatic potential of cancer cells.17 Thus, Vimentin has gained much attention as a canonical tumor marker. As a result, discovering the relationship between circRNAs and Vimentin will provide novel insights for OS treatment.
Materials and Methods
Ethical Approval
All animal experiments were approved by the Ethics Committee of Sir Run Run Shaw Hospital and carried out under the guidelines of the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health.
Patients and Tissue Collection
From April 2018 to April 2019, ten primary osteosarcoma and ten chondroma patients who underwent radical surgery at the Sir Run Run Shaw Hospital, Zhejiang, China, were included in this study. This study was approved by the Ethics Review Committees of Sir Run Run Shaw Hospital in accordance with the Declaration of Helsinki, and informed consents were signed by the patients prior to using the clinical samples. All the resected specimens were placed into liquid nitrogen immediately and stored at −80°C. All the patients had received the same chemotherapy regimen before surgery.
Cell Culture and Cell Transfection
Four human osteosarcoma cell lines (143B, HOS, U20S, and MG-63), HEK-293 and human osteoblast cells hFOB1.19 were commercially acquired from the Chinese Academy of Sciences (Shanghai, China). Osteosarcoma cell lines and HEK-293 were cultured in DMEM supplemented with 10% FBS (Gibco, Gran Island, NY, USA), 100 U/mL penicillin, and 100 U/mL streptomycin (Invitrogen, Carlsbad, CA, USA), while hFOB1.19 cells were maintained in Ham’s F12/ DMEM supplemented with 10% FBS, 100 U/mL penicillin and 100 mg/mL streptomycin. All the cells were incubated at 37°C with 5% CO2.
According to the information of hsa_circ_0007031 (circTUBGCP3) from GenBank, the sequences of the short siRNAs (GCAATAATGTGGTCTACAA) targeting spliced junction of circTUBGCP3 and negative control-siRNA (si-NC) were constructed by RiboBio (Guangzhou, China). The synthetic siRNA sequences were subcloned into the pcDNA3.1 vector (Invitrogen). hsa_circ_0007031 down-regulation was achieved through pcDNA3.1-siRNA transfection using Lipofectamine 3000 reagent according to the manufacturer’s instructions. The surviving cells were continuously cultured as stable mass transfectants.
RNA Isolation
Total RNA was isolated from cells, tissues, or serum samples using the TRIzol kit (Invitrogen, Carlsbad, CA, USA) following the manufacture’s guide and quantified using a NanoDrop ND-1000 spectrophotometer at 260 nm (NanoDrop, Wilmington, DE, USA).
qRT-PCR
To detect the expression of hsa_circ_0007031 (circTUBGCP3) in OS cell lines and clinical samples, quantitative real-time PCR (qRT-PCR) analysis was performed using SYBR green kit (TaKaRa, Dalian, China) on a Roche LightCycler® 480II PCR machine (Basel, Switzerland) in accordance with the manufacturer’s protocols. GAPDH was used for normalizing the expression of circTUBGCP3. The corresponding expression data was measured and relative expression was calculated by using the equation 2-ΔΔCT method. Primers used for amplifying hsa_circ_0007031 in this study were designed as divergent primers cross the splicing site (F: 5’AGGCCCAGGATTTGGATCAC3’ R: 5’CTTGTCGTGCCACAGTCGAT3’).
Cell Counting Kit-8 Assay
CCK-8 assay was performed to measuring cell vitality of OS cells using CCK-8 kit (Abmole Bioscience, Shanghai, China). After 24 h, transfected cells at the logarithmic growth stage were seeded into 96-well plates at a density of 5 × 104 cells in a total volume of 100 µL per well. Once the cells adhered to the surface, 10 µL CCK-8/well was added and incubated for 24, 48, 72 and 96h. The absorbance value of each well was measured with a spectrophotometer set at 450 nm.
Apoptosis Assay
Detection of apoptosis was carried out by flow cytometric assays with Annexin V-FITC/PI cell apoptosis detection kit (BioGems). Transfected cells in logarithmic growth phase were washed, collected, and resuspended in the binding buffer. The cells were then stained with Annexin V-FITC and PI and co-incubated in darkness for 15 min. Ultimately, the samples were subjected to flow cytometry based apoptosis analysis within an hour.
Migration Assay
The migration assay was performed using Transwell chambers according to the manufacturer’s instructions. Transfected cells in logarithmic growth phase were washed twice with PBS and 104 cells resuspended in 250 μL serum-free DMEM were added into the upper chamber of the Transwell insert. The lower chamber consisted of complete DMEM. After 24h incubation at 37 °C, the media and the non-invading cells in the upper chamber were removed with a cotton swab. The migrated cells were fixed with 4% paraformaldehyde solution and subsequently stained with 10% crystal violet. The observation fields of vision were chosen randomly using a Leica inverted microscope.
Luciferase Reporter Assays
The sequence of circTUBGCP3 and its corresponding mutants were cloned into the pmirGLO luciferase vector. OS cells were incubated at around 60% confluency and subsequently seeded in 6-well plates. They were then cotransfected with wild type or mutant luciferase reporter vector and miR-30b mimic or negative control mimic (mimic NC). Cell extracts were prepared 48 h after transfection and luciferase activity was measured using dual-luciferase reporter assay system. The fold change in luciferase activity was calculated in the corresponding miRNA co-transfected cells as compared with NC.
Subcutaneous Orthotopic Xenograft Tumor Models
Nude mice (male, 4 weeks old) were injected subcutaneously with a total of 5 × 106 143B stable cells. Tumor volume was calculated according to the length (a) and the width (b) by using the following formula: volume (mm3)=a b2/2. Four weeks after injection, the animals were sacrificed, and tumors were harvested and fixed in 4% paraformaldehyde. Wet tumor weight was calculated as mean weight ± standard deviation (SD) in each group.
RNA Immunoprecipitation
RIP experiments were performed by using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). HEK-293 cells were transfected with the Ago2 vector or plasmid. Approximately 1×107 HOS cells were pelleted and re-suspended with an equal pellet volume of RIP Lysis Buffer (about 100 mL) combined with protease inhibitors cocktail and RNase inhibitors. The cell lysates (200 μL) were incubated with 5 μg of antibody against Ago2 (Millipore) or rabbit IgG-coated beads and rotated at 4 °C overnight. After treating the lysates with proteinase K buffer, immunoprecipitated RNA was extracted by using the RNeasy MinElute Cleanup Kit (Qiagen) and reverse transcribed using Prime-Script RT Master Mix (TaKaRa). The abundance of circTUBGCP3 was detected by qRT-PCR.
Pull-Down Assay with Biotinylated circTUBGCP3 Probe
Pull-down assay was performed as indicated: Briefly, 1× 107 osteosarcoma cells were harvested, lysed, and sonicated. The circTUBGCP3 probe was incubated with C-1 magnetic beads (Life Technologies) at 25°C for 2 h to generate probe-coated beads. The cell lysates were incubated with circTUBGCP3 probe or oligo probe at 4°C overnight. After washing with the wash buffer, the RNA complexes bound to the beads were eluted and extracted with RNeasy Mini Kit (QIAGEN) for RT–PCR or qRT PCR. Biotinylated circTUBGCP3 probe was designed and synthesized by RiboBio (Guangzhou, China).
Statistical Analysis
All the experiments were performed as three independent experiments in triplicates. Data analyses were conducted using Graph Pad Prism 5.01 (SanDiego, CA). Error bars represent standard deviation (SD) of a triplicate set of experiments. Quantitative data were expressed as mean ± SD. Significant differences for quantitative data were compared by Student’s t-test. P < 0.05 was considered significant.
Results
circTUBGCP3 Is Highly Expressed in OS Tissues and Cell Lines
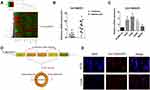
A microarray expression profile comparing circRNAs in OS cell lines with those in hFOB1.19 cells has been described previously (GSE96964)18. We discovered that the expression level of circTUBGCP3 was remarkably enhanced in multiple OS cell lines compared with hFOB1.19 cells, a normal osteoblast cell line (Figure 1A). To determine the correlation between circTUBGCP3 expression and OS, 10 pairs of chondroma and OS tissue samples were selected, and subjected to qRT-PCR based detection of the expression of circTUBGCP3. Our results verified the relative abundance of circTUBGCP3 between OS and chondroma tissue (Figure 1B). According to the relative mRNA expression, the expression of circTUBGCP3 was predominately higher in multiple OS cell lines (HOS, 143B, U2OS, and MG63) than in the hFOB1.19 cell line. Among the OS cell lines, HOS and 143B cells demonstrated the highest levels of circTUBGCP3 as compared to other cells (Figure 1C). We further compared TUBGCP3 mRNA sequences with the expected sequences of circTUBGCP3 acquired from circBase and verified that circTUBGCP3 was looped comprising the exons 12 and 19 of its parental gene. We further confirmed the head-to-tail splicing site of circTUBGCP3 via Sanger sequencing (Figure 1D). Furthermore, RNA FISH exhibited that circTUBGCP3 was mostly localized in the cytoplasm (Figure 1E).
Down-Regulating circTUBGCP3 Inhibits the Proliferation, Migration and Survivability of OS Cells
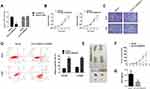
To knock down the expression of circTUBGCP3 in the cells stably, circTUBGCP3 small hairpin RNAs (shRNAs) were constructed to investigate the function of circTUBGCP3 in OS cells. One sh-circTUBGCP3 was designed specifically to target the junction sites of this circRNA and transfected them into 143B and HOS cells. Satisfactory transfection efficiency was achieved 48h post-transfection which was identified by qPCR. As shown in Figure 2A, the expression of circTUBGCP3 was significantly reduced in OS cells, while the expression of TUBGCP3 mRNA did not change. CCK-8 assay was performed to detect the vitality of OS cell in presence or absence of circTUBGCP3, which demonstrated the crucial role of circTUBGCP3 in OS cell proliferation (Figure 2B). To explore whether circTUBGCP3 was involved in OS cell migration, transwell migration assays were performed. The results indicated that the motility of OS cells was evidently reduced upon silencing circTUBGCP3 using shRNA (Figure 2C). In addition, the results of apoptosis assay suggest that the knockdown of circTUBGCP3 expression prominently enhances the apoptosis in OS cells (Figure 2D). To investigate the impact of circTUBGCP3 in vivo, circTUBGCP3 knockdown or control 143B cells were subcutaneously injected into nude mice and tumorigenicity was assessed. The results revealed that the cells lacking circTUBGCP3 exhibited a reduced growth rate as compared with the control cells (Figure 2E–G).
circTUBGCP3 Functions as a miR-30b Sponge in OS Cells
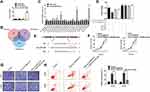
Various studies have demonstrated that circRNAs could bind to miRNA and function as miRNA sponges and partially hinder the function of the respective miRNAs. The Ago-RIP assay was conducted in HOS cells stably expressing vector or shcircTUBGCP3, and we found that the endogenous circTUBGCP3 pulled down by anti-AGO2 antibodies was notably enriched in the AGO2 overexpression group by qRT–PCR compared with the control group, indicating that circTUBGCP3 could bind and interact with miRNAs through AGO2 protein (Figure 3A). Three databases (miRanda, TargetScan, and RNAhybrid) were used to predict the potential target miRNAs for circTUBGCP3, and 22 candidate miRNAs were selected from the overlap between the databases (Figure 3B). Next, to confirm whether circTUBGCP3 could sponge candidate miRNAs, we conducted a pull-down assay with a biotin-labeled circTUBGCP3 probe in OS cells, and 3 microRNAs were pulled down by circTUBGCP3 in HOS cells (Figure 3C). We further confirmed the binding of 3 miRNAs by the results of the luciferase assay (Figure 3D). We mutated this response element in the 3′-untranslated region (3′-UTR) of circTUBGCP3 and cloned it into a luciferase reporter vector. Three miRNAs (miR-30b, miR-1266 and miR-6812) were co-transfected with mutant (Mut) or wild-type (WT) 3′ UTR of circTUBGCP3 into HEK-293 cells, and luciferase activity was measured. The results revealed that the luciferase activities of the Mut reporter had obviously higher luciferase activity than the wild-type (WT) reporter (Figure 3D).The schematic diagram of the binding sites for miR-30b in circTUBGCP3 are represented in Figure 3E. In silico analysis of Circular RNA Interactome suggests that circTUBGCP3 may bind to miR-30b. Moreover, circTUBGCP3 is identified to possess a miRNA response element for miRNA-30b. Rescue assays were further performed by co-transfecting sh-circTUBGCP3 with miR-30b sponge to verify that circTUBGCP3 functions as a miR-30b sponge. The result of CCK-8 and Flow cytometric based apoptosis analysis as well as transwell migration assays prove that miR-30b sponge partially rescues the suppression of OS cell proliferation caused by sh-circTUBGCP3 (Figure 3F–H).
circTUBGCP3 Facilitates OS Progression via Vimentin
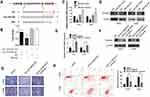
After identifying the circTUBGCP3 role as a miR-30b sponge, we further investigated whether circTUBGCP3 can promote OS cell progression via Vimentin. To examine whether miR-30b could directly target Vimentin, we cloned the Vimentin mRNA into the luciferase vector and subsequently co-transfected the same in HEK-293 cells with miR-30b mimics. With the overexpression of miR-30b, the luciferase activity of the Vimentin luciferase was found to be reduced as compared to the control group. Furthermore, a mutated form of the Vimentin 3′ UTR was generated by interfering with the binding sequence of miR-30b and co-transfected with miR-30b mimics. The results of subsequent assessment of luciferase activity revealed that the cells transfected with Mutant Vimentin 3′ UTR showed higher luciferase activity as compared to those transfected with wild-type Vimentin 3′ UTR (Figure 4A and B). These data suggest that Vimentin might be a trigger gene in OS, and miR-30b directly interacts with Vimentin 3′ UTR. To verify that miR-30b targets Vimentin, qPCR and Western blot were performed. As a result, HOS and 143B cells co-transfected with miR-30b mimic showed decreased Vimentin expression at the level of mRNA and protein, while those co-transfected with miR-30b inhibitor showed enhanced Vimentin expression in the level of mRNA and protein (Figure 4C and D). We subsequently performed rescue assays to detect the protein and mRNA level of Vimentin by co-transfecting sh-circTUBGCP3 with Vimentin. The results showed that the expression of Vimentin at the level of mRNA and protein was enhanced after rescue assay (Figure 4E and F). Further, Vimentin could moderately rescue the OS cell suppression due to sh-circTUBGCP3, which was demonstrated by transwell migration assay and flow cytometric based apoptosis analysis (Figure 4G and H).
Discussion
CircRNAs are a subclass of novel long non-coding RNA with an uncommon circular structure.8 Recent studies have revealed that circRNAs overexpress in multiple cancers and positively correlate with cancer progression.19–21 Thus, they provide researchers with a new avenue for exploring their role in identifying the cancer prognosis. Several circRNAs can serve as clinically relevant biomarkers for the diagnosis and prognosis of cancers. For instance, Hansen et al22 have shown that ciRS-7 is the key pathogenic factor of many neurological and brain tumor diseases. Wang et al23 have reported that hsa_circ_001988 is down-regulated in colorectal cancer tissues and may serve as a novel potential biomarker in the diagnosis and treatment of colorectal cancer. Additionally, circRNAs could inhibit the function of miRNA through sponge action and further alter the expression of downstream target genes to perform biological functions.24 Zhu et al25 have found that circ-0013958 functions as a scaffold to inhibit miRNA-134 and removes the inhibition of miRNA-134 on Cyclin D1 in cancer cells. Although there has been tremendous progress in circRNAs research, the progress in the understanding towards concrete functional mechanism and possible clinical significance between circTUBGCP3 and OS are still limited.
The TUBGCP3 (Tubulin Gamma Complex Associated Protein 3) is an important part of Gamma-tubulin complex, which is necessary for microtubule nucleation at the centrosome.26 Previous studies have reported that TUBGCP3 plays a vital part in forming the structural constituent of cytoskeleton and microtubule nucleation by interphase microtubule organizing center.27 It is well established that circRNAs originate from a series of exons. By Sanger sequencing, we have further confirmed that circTUBGCP3 is derived from exons 12 and 19 of TUBGCP3 gene and constitutes a special loop structure by head-to-tail splicing. In the current study, ten paired groups of OS tissues obtained from Sir Run Run Shaw Hospital were subjected to qPCR assays which demonstrated the up-regulation of circTUBGCP3 expression level separately in four kind of OS cells lines in comparison to the normal osteoblast cells (hFOB1.19). Additionally, the expression level of circTUBGCP3 is evidently up-regulated in OS samples in contrast to the chondroma samples suggesting that circTUBGCP3 might play an essential role in the progression of OS. Besides, a series of functional assays further indicated that overexpression of circTUBGCP3 serves as an oncogenic role in OS, promoting cell proliferation, survivability, and migration. In addition, dual-luciferase reporter assays were conducted to further identify the mechanism of circTUBGCP3 in OS cells. The result revealed that circTUBGCP3 sponges and negatively regulates miR-30b. Furthermore, we have corroborated our in vitro findings in nude mice by performing tumorigenicity assay in vivo. To sum up, our research indicates that circTUBGCP3 regulates the Vimentin mediated OS progression by sponging miR-30b. Vimentin is an intermediate filament protein. In multiple epithelial cancers, such as prostate cancer, gastrointestinal tumors, breast cancer and so on, the expression level of Vimentin is found to be increased.28 In this study, our research demonstrates that Vimentin is a trigger gene in OS through a variety of rescue assays. Thus, by means of sponging miR-30b that targets Vimentin, circTUBGCP3 accelerates the proliferation, migration and survivability of OS cells.
Conclusion
In summary, we have demonstrated that the expression level of circTUBGCP3 is remarkably up regulated in OS tissue specimens and cells in comparison with the normal tissues and cells. Furthermore, down regulation of circTUBGCP3 significantly promotes OS cell proliferation, migration, and survivability. Thus, our findings potentially accelerate the research of circTUBGCP3 as a potential target in OS therapy.
Abbreviations
OS, osteosarcoma; circ, circular RNA; qRT-PCR, quantitative real-time PCR; TUBGCP3, Tubulin Gamma Complex Associated Protein 3.
Acknowledgments
The research was supported by the National Natural Science Foundation of China (grant number 81972504), the Medical Science and Technology Project of Zhejiang Province of China (grant number 2020388803), the Zhejiang Provincial Natural Science Foundation of China (grant number LY19H160058), and the Zhejiang Provincial Natural Science Foundation of China (grant number LY16G030006).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhang Y, Cheng H, Li W, et al. Highly-expressed P2X7 receptor promotes growth and metastasis of human HOS/MNNG osteosarcoma cells via PI3K/Akt/GSK3β/β-catenin and mTOR/HIF1α/VEGF signaling. Int j Cancer. 2019;145(4):1068–1082. doi:10.1002/ijc.32207
2. Kim M, Jung JY, Choi S, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13(1):149–168. doi:10.1080/15548627.2016.1239676
3. Ji T, Guo Y, Kim K, et al. Neuropilin-2 expression is inhibited by secreted Wnt antagonists and its down-regulation is associated with reduced tumor growth and metastasis in osteosarcoma. Mol Cancer. 2015;14(1):86. doi:10.1186/s12943-015-0359-4
4. Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–790. doi:10.1200/JCO.2002.20.3.776
5. Angulo P, Kaushik G, Subramaniam D, et al. Natural compounds targeting major cell signaling pathways: a novel paradigm for osteosarcoma therapy. J Hematol Oncol. 2017;10(1):10. doi:10.1186/s13045-016-0373-z
6. Hou CH, Lin FL, Hou SM, et al. Cyr61 promotes epithelial-mesenchymal transition and tumor metastasis of osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol Cancer. 2014;13(1):236. doi:10.1186/1476-4598-13-236
7. Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559
8. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi:10.1016/j.molcel.2013.08.017
9. Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234(1):500–508. doi:10.1002/jcp.26612
10. Huang L, Chen M, Pan J, et al. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 2018;500(2):511–517. doi:10.1016/j.bbrc.2018.04.131
11. Nie WB, Zhao LM, Guo R, et al. Circular RNA circ-NT5C2 acts as a potential novel biomarker for prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(19):6239–6244. doi:10.26355/eurrev_201810_16030
12. Du YX, Guo LX, Pan HS, et al. Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion. Hum Cell. 2019;33:252–260.
13. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi:10.7150/jca.24619
14. Messica Y, Laser-Azogui A, Volberg T, et al. The role of vimentin in regulating cell invasive migration in dense cultures of breast carcinoma cells. Nano Lett. 2017;17(11):6941–6948. doi:10.1021/acs.nanolett.7b03358
15. Burikhanov R, Sviripa VM, Hebbar N, et al. Arylquins target vimentin to trigger Par-4 secretion for tumor cell apoptosis. Nat Chem Biol. 2014;10(11):924–926. doi:10.1038/nchembio.1631
16. McDonald-Hyman C, Muller JT, Loschi M, et al. The vimentin intermediate filament network restrains regulatory T cell suppression of graft-versus-host disease. J Clin Invest. 2018;128(10):4604–4621. doi:10.1172/JCI95713
17. Zhu Z, Rong Z, Luo Z, et al. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer. 2019;18(1):126. doi:10.1186/s12943-019-1054-7
18. Liu W, Zhang J, Zou C, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43(3):969–985. doi:10.1159/000481650
19. Wang Y, Li Y, He H, et al. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi:10.1016/j.gene.2019.144099
20. Ding L, Zhao Y, Dang S, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45. doi:10.1186/s12943-019-1006-2
21. Bi J, Liu H, Dong W, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18(1):133. doi:10.1186/s12943-019-1060-9
22. Hollensen AK, Andersen S, Hjorth K, et al. Enhanced tailored microRNA sponge activity of RNA Pol II-transcribed TuD hairpins relative to ectopically expressed ciRS7-derived circRNAs. Mol Ther Nucleic Acids. 2018;13:365–375. doi:10.1016/j.omtn.2018.09.009
23. Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025.
24. Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi:10.1038/cdd.2017.86
25. Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi:10.1111/febs.14132
26. Li G, Jin D, Zhong TP. Tubgcp3 is required for retinal progenitor cell proliferation during zebrafish development. Front Mol Neurosci. 2019;12:126. doi:10.3389/fnmol.2019.00126
27. Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–3239. doi:10.2337/db10-0502
28. Colburn ZT, Jones JCR. Complexes of alpha6beta4 integrin and vimentin act as signaling hubs to regulate epithelial cell migration. J Cell Sci. 2018;131:jcs214593.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.