Back to Journals » Cancer Management and Research » Volume 12
Circular RNA circRNA_103809 Accelerates Bladder Cancer Progression and Enhances Chemo-Resistance by Activation of miR-516a-5p/FBXL18 Axis
Authors Huang W, Lu Y, Wang F, Huang X, Yu Z
Received 16 May 2020
Accepted for publication 29 July 2020
Published 21 August 2020 Volume 2020:12 Pages 7561—7568
DOI https://doi.org/10.2147/CMAR.S263083
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Weiping Huang, Yongyong Lu, Feng Wang, Xixi Huang, Zhixian Yu
Department of Urology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Correspondence: Zhixian Yu
Department of Urology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325000, People’s Republic of China
Email [email protected]
Background: Numerous researches have suggested that circular RNAs (circRNAs) play critical functions in bladder cancer (BC) progression. This study aims to investigate the potential roles of circRNA_103809 in regulating BC development.
Methods: qRT-PCR was used to analyze gene expression. CCK8 and colony formation were used to analyze cell proliferation. Transwell was utilized to examine cell migration and invasion. Gemcitabine was used to analyze the effect of circRNA_103809 on the chemo-resistance of BC cells. Luciferase reporter assay was performed to detect the RNA interactions.
Results: circRNA_103809 was highly expressed in BC tissues and cell lines. CircRNA_103809 high expression was associated with a poor progression in BC patients. CircRNA_103809 knockdown impaired the growth and metastasis of BC cells. Furthermore, circRNA_103809 silencing increased the sensitivity of BC cells to Gemcitabine treatment. CircRNA_103809 was the sponge for miR-516a-5p and promoted FBXL18 expression via restraining miR-516a-5p activity.
Conclusion: circRNA_103809 promotes proliferation, migration, invasion and chemo-resistance of BC cells through regulating miR-516a-5p/FBXL18 axis.
Keywords: bladder cancer, circRNA_103809, miR-516a-5p, FBXL18
Introduction
Bladder cancer (BC) derives from the urinary system and has a high incidence.1 Due to its malignance, BC gives rise to a large amount of cancer-related deaths every year around the world.2 Due to high rates of recurrence and metastasis, BC patients display a quite poor prognosis. Notably, treatment using chemotherapy or radiotherapy on advanced BC shows limited benefit. Thus, it is required to develop novel therapeutic targets to BC intervention.
Circular RNAs (circRNAs) are a novel member of noncoding RNAs (ncRNAs) and characterized by a closed-loop structure and lacking coding potential.3 As the advance of high-throughput sequencing method, circRNAs have been demonstrated to be widely expressed in various tissues and cells.4 Importantly, researches have proven that circRNA dysregulation is associated with multiple human diseases, such as neurological disorder and carcinomas.5,6 Many circRNAs have miRNA binding elements and work as competing endogenous RNAs (ceRNAs) to inhibit miRNA and promote gene expression.7 And circRNAs could participate in tumorigenesis through regulating several biological processes, including cell proliferation and metastasis.8 For instance, knockdown of hsa_circRNA_002178 suppresses breast cancer development through sponging miR-328-3p to upregulate COL1A1expression.9 CircRNA_104433 promotes gastric cancer cell growth and migration via regulating miR-497-5p.10 Thus, circRNA-miRNA-mRNA signaling cascade plays an important role in cancer progression.
CircRNA_103809 has been reported to promote lung cancer development.11 Besides, other researches showed that circRNA_103809 initiates tumorigenesis in liver and colorectum.12,13 This study aimed to investigate the roles of circRNA_103809 in BC and illustrate its functional mechanism.
Materials and Methods
Tissue Collection
55 BC tissues and corresponding adjacent normal tissues were collected from the First Affiliated Hospital of Wenzhou Medical University. This study was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University (No. 20,180,418,126). We achieved written informed consent from the involved patient. All tissues were not treated with chemotherapy or radiotherapy before surgery and stored in liquid nitrogen until use.
Cell Culture and Transfection
BC cell lines and human immortalized uroepithelium cell line SV-HUC-1 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (GIBCO BRL, NY, USA) and maintained in a humidified atmosphere with 5% CO2. Small interfering RNA (siRNA) targeting circRNA_103809, miR-516a-5p mimics, miR-516a-5p inhibitors and corresponding negative controls were purchased from Gene-Pharma (Shanghai, China). Cell transfection was carried out by using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions.
qRT-PCR
Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions. Then, RNA was reversely transcribed into complementary DNA (cDNA) using PrimeScript RT Master Mix (Takara, Dalian, China) followed by qPCR examination using PCR Master Mix (2×) (ThermoFisher Scientific, Waltham, Ma, USA). Relative expression was normalized to GAPDH or U6 and calculated according to the 2−∆∆CT method.
CCK8 Assay
Cells were seeded into the 96-well plates and cultured for the indicated time. Then, CCK8 solution was added and incubated for 2 h. The absorbance at 450 nm was detected with an automatic enzyme-linked immune detector.
Colony Formation Assay
T24 and EJ cells were seeded into the 6-well plate and incubated for 14 days. Then, colonies were fixed with 4% paraformaldehyde and dyed with crystal violet.
Transwell Assay
To examine migration and invasion, cells were seeded into a 24-well transwell chamber (Costar, USA) with or without precoated Matrigel. Cells in the upper chamber were cultured using 200 μL serum-free medium. The lower chamber was fixed with 10% FBS-supplemented medium. After incubating for 24 h, the cells that migrated or invaded into the lower membrane surface were fixed with 4% paraformaldehyde and stained with 1% crystal violet. Cell numbers were finally counted.
Luciferase Reporter Assay
The binding site between circRNA_103809 and miR-516a-5p was predicted using Circinteractome (https://circinteractome.nia.nih.gov/). The binding site between miR-516a-5p and FBXL18 was predicted using TargetScan7 (http://www.targetscan.org/vert_71/) and miRDB (http://mirdb.org/miRDB/index.html). To perform the luciferase reporter assay, the wild-type or mutant sequence of circRNA_103809 and FBXL18 3ʹUTR was inserted into pGL luciferase vector (Promega, USA). Then, luciferase vector and miR-516a-5p mimics were transfected into EJ cells. After 48 h, the luciferase activity was measured using a dual-luciferase reporter assay system (Promega, USA) according to the manufacturer’s protocol. Renilla luciferase activity was normalized control.
Statistical Analysis
Statistical analyses were performed by GraphPad Prism 7.0 software. Data are presented as the mean ± SD. Survival rate was analyzed by Kaplan-Meier method and Log rank test. Student’s t-test or one-way ANOVA followed by Tukey’s post hoc test was used for the analysis of statistically significant differences. P < 0.05 was considered statistically significant.
Results
Expression Pattern of circRNA_103809 in BC
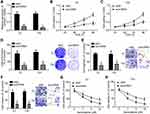
55 BC tissues and their adjacent normal tissues were collected. Then, circRNA_103809 expression was firstly examined. It was found that circRNA_103809 expression was upregulated in BC tissues (Figure 1A). Besides, it was observed that circRNA_103809 expression was also elevated in BC cell lines compared to SV-HUC-1 cells (Figure 1B). According to circRNA_103809 median value, BC tissues were divided into low expression group and high expression group. After analysis, we found that circRNA_103809 high expression was inversely correlated with overall and progression-free survival rates (Figure 1C and D).
Effects of circRNA_103809 Knockdown on BC Progression and Chemo-Resistance
EJ and T24 cells were selected for experiments. After transfection with sicircRNA_103809, its expression was successfully decreased (Figure 2A). CCK8 assay showed that circRNA_103809 depletion inhibited the proliferation of EJ and T24 cells (Figure 2B and C). Colony formation result further validated the result that circRNA_103809 knockdown inhibited proliferation (Figure 2D). Transwell assay was then conducted. CircRNA_103809 knockdown decreased the cell numbers of migration and invasion (Figure 2E and F). Notably, we found that circRNA_103809 knockdown decreased the resistance of EJ and T24 cells to Gemcitabine treatment (Figure 2G and H).
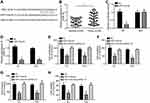
circRNA_103809 Was the ceRNA for miR-516a-5p
Then, the potential target of circRNA_103809 was analyzed by bioinformatics. We found miR-516a-5p and constructed wild-type (Wt) and mutant (Mut) luciferase reporters (Figure 3A). We noticed that miR-516a-5p expression was downregulated in BC tissues (Figure 3B). And its expression was inversely associated with circRNA_103809 level in BC tissues (Figure 3C). Luciferase reporter assay was performed. Results illustrated that circRNA_103809-Wt luciferase activity was repressed by miR-516a-5p mimics (Figure 3D). And miR-516a-5p expression was increased after circRNA_103809 knockdown (Figure 3E). Thus, circRNA_103809 was a ceRNA for miR-516a-5p.
miR-516a-5p Directly Targeted FBXL18
Afterwards, the potential target of miR-516a-5p was analyzed by bioinformatics. We further identified FBXL18 (F-box and leucine-rich repeat protein 18), a regulator of apoptosis and cell cycle progression.14 We then generated corresponding luciferase reporter vectors (Figure 4A). FBXL18 expression was increased in BC tissues compared to normal controls (Figure 4B). Luciferase reporter assay demonstrated the direct interaction between miR-516a-5p and FBXL18 (Figure 4C). Importantly, miR-516a-5p inhibited the level of FBXL18 in EJ and T24 cells (Figure 4D). To explore the roles of miR-516a-5p and FBXL18 in BC, we performed CCK8 and Transwell assays. We found that miR-516a-5p mimics inhibited the proliferation, migration, invasion and resistance to Gemcitabine in BC cells (Figure 4E–H).
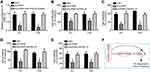
circRNA_103809 Contributed to the Malignant Behaviors of BC Cells Through miR-516a-5p/FBXL18 Axis
Interestingly, it was observed that circRNA_103809 knockdown inhibited FBXL18 expression while miR-516a-5p inhibitors abrogated this trend (Figure 5A). We further sought to determine the role of FBXL18 in circRNA_103809-mediated BC progression. Through CCK8 and transwell assays, we discovered that restoration of FBXL18 expression rescued the abilities of proliferation, migration, invasion and chemo-resistance in BC cells transfection with sicircRNA_103809 (Figure 5B–E). Summarily, circRNA_103809 enhances the malignant behaviors of BC cells via regulating miR-516a-5p/FBXL18 axis (Figure 5F).
Discussion
There is an urgent requirement to understand the molecular mechanism underlying BC development. In this study, we showed that circRNA_103809 expression was upregulated in BC tissues. And circRNA_103809 could be a potential prognostic biomarker for BC patients. Functional experiments showed that circRNA_103809 knockdown repressed BC cell proliferation, metastasis and chemo-resistance. Moreover, mechanistic results identified that circRNA_103809 was a ceRNA for miR-516a-5p to promote FBXL18 expression. Our findings highlighted the crucial roles of circRNA_103809/miR-516a-5p/FBXL18 axis in BC progression.
So many references have demonstrated the key functions of circRNAs in cancers, including BC.8,15 For instance, circMYLK affects the proliferation and metastasis of BC cells by regulating VEGFA/VEGFR2 pathway.15 Circ-CEP128 upregulation in BC promotes tumor cell growth, migration and invasion through sponging miR-145 to upregulating SOX11 expression.16 Liu et al firstly reported that circRNA_103809 contributes to lung cancer cell proliferation, migration and invasion.11 Then, Bian et al indicated that circRNA_103809 is involved in the regulation of cell proliferation, migration and invasion in colorectal cancer.12 Additionally, Zhan et al found that circRNA_103809 enhances growth and invasiveness of liver cancer.13 In our study, we revealed that circRNA_103809 expression was significantly upregulated in BC tissues and cell lines. Moreover, through CCK8, colony formation and transwell assays, we demonstrated that circRNA_103809 downregulation repressed the growth, metastasis and chemo-resistance of BC cells. Our study proved that circRNA_103809 is an important regulator of BC progression.
Recently, researches have suggested that circRNAs are potential sponges for miRNAs by direct interaction.8,11 For example, circRNA circUBXN7 suppresses BC cell proliferation and migration through sponging miR-1247 to upregulate B4GALT3 level.17 CircRNA-CEP128 interacts with miR-145-5p to enhance Myd88 expression and contribute to BC development.18 In addition, circRNA hsa_circ_0068871 promotes BC tumorigenesis through inhibiting miR-181a-5p expression to facilitate the expression of FGFR3 and activation of STAT3.19 circRNA_103809 was also identified to sponge several miRNAs, such as miR-4302 and miR-532-3p.11,12 In our study, we identified that circRNA_103809 was the sponge for another miRNA miR-516a-5p. We proved their interaction through luciferase reporter assay and demonstrated their regulatory relationship. MiR-516a-5p is a tumor suppressor in hepatocellular carcinoma and lung cancer.20,21 Whether miR-516a-5p affects BC development is unknown. In our study, we found that miR-516a-5p mimics suppressed the growth, metastasis and chemo-resistance in BC. Thus, miR-516a-5p is a new tumor suppressor for BC.
Afterwards, we identified that FBXL18 is a direct target of miR-516a-5p. And we further demonstrated that FBXL18 expression was regulated by circRNA_103809/miR-516a-5p axis. Only one study defined the role of FBXL18 in cancer.22 Zhang et al showed FBXL18 promotes glioma proliferation.22 Our findings showed that FBXL18 was upregulated in BC tissues. And FBXL18 restoration abolished the anti-tumor effects of circRNA_103809 knockdown or miR-516a-5p mimics in BC. Thus, our findings verified that FBXL18 is a downstream molecule of circRNA_103809/miR-516a-5p axis and promotes BC progression.
In summary, the current results demonstrate that circRNA_103809 overexpression in BC promotes tumor progression and enhances chemo-resistance through targeting miR-516a-5p/FBXL18 signaling. This finding indicates that circRNA_103809 may be a novel therapeutic target.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Li Y, Zheng F, Xiao X, et al. Circ HIPK 3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi:10.15252/embr.201643581
3. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
4. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO, Moran JV. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi:10.1371/journal.pgen.1003777
5. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
6. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. doi:10.1038/ncomms11215
7. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
8. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508(4):991–996. doi:10.1016/j.bbrc.2018.12.046
9. Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J Cell Mol Med. 2020;24(3):2189–2201. doi:10.1111/jcmm.14875
10. Wei W, Mo X, Yan L, et al. Circular RNA profiling reveals that circRNA_104433 regulates cell growth by targeting miR-497-5p in gastric cancer. Cancer Manag Res. 2020;12:15–30. doi:10.2147/CMAR.S219307
11. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
12. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352. doi:10.1016/j.bbrc.2018.09.073
13. Zhan W, Liao X, Chen Z, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235(2):1733–1745. doi:10.1002/jcp.29092
14. Liu Y, Lear T, Zhao Y, et al. F-box protein Fbxl18 mediates polyubiquitylation and proteasomal degradation of the pro-apoptotic SCF subunit Fbxl7. Cell Death Dis. 2015;6(2):e1630. doi:10.1038/cddis.2014.585
15. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027
16. Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24(1):40. doi:10.1186/s10020-018-0039-0
17. Liu H, Chen D, Bi J, et al. Circular RNA circUBXN7 represses cell growth and invasion by sponging miR-1247-3p to enhance B4GALT3 expression in bladder cancer. Aging (Albany NY). 2018;10(10):2606–2623. doi:10.18632/aging.101573
18. Sun M, Zhao W, Chen Z, et al. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer. 2019;145(8):2170–2181. doi:10.1002/ijc.32311
19. Mao W, Huang X, Wang L, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38(1):169. doi:10.1186/s13046-019-1136-9
20. Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi:10.1038/s41419-019-2176-y
21. Ye XY, Xu L, Lu S, Chen ZW. MiR-516a-5p inhibits the proliferation of non-small cell lung cancer by targeting HIST3H2A. Int J Immunopathol Pharmacol. 2019;33:2058738419841481. doi:10.1177/2058738419841481
22. Zhang J, Yang Z, Ou J, Xia X, Zhi F, Cui J. The F-box protein FBXL18 promotes glioma progression by promoting K63-linked ubiquitination of Akt. FEBS Lett. 2017;591(1):145–154. doi:10.1002/1873-3468.12521
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.