Back to Journals » Cancer Management and Research » Volume 12
Circular RNA CircPRMT5 Accelerates Proliferation and Invasion of Papillary Thyroid Cancer Through Regulation of miR-30c/E2F3 Axis
Authors Xue C, Cheng Y, Wu J, Ke K, Miao C, Chen E, Zhang L
Received 11 February 2020
Accepted for publication 21 April 2020
Published 11 May 2020 Volume 2020:12 Pages 3285—3291
DOI https://doi.org/10.2147/CMAR.S249237
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Cheng Xue,1 Yi Cheng,1 Jinyou Wu,1 Kongliang Ke,2 Chundi Miao,2 Enfu Chen,1 Luqing Zhang2
1Department of Endocrinology, Wenling First People’s Hospital, Wenling 317500, People’s Republic of China; 2Department of Anorectal Surgery, Ningbo Hangzhou Bay Hospital, Ningbo 315000, People’s Republic of China
Correspondence: Luqing Zhang
Department of Anorectal Surgery, Ningbo Hangzhou Bay Hospital, 1155 Binhai 2nd Road, Hangzhouwan New District, Ningbo 315000, People’s Republic of China
Email [email protected]
Background: The role of circular RNA (circRNA) in papillary thyroid cancer (PTC) is largely unknown. This study aims to determine the function and mechanism of circPRMT5 in the regulation of PTC development.
Methods: PTC tissues and cell lines were used to determine circPRMT5 expression via quantitative real-time polymerase chain reaction. Small interfering RNA (siRNA) was utilized to knock down circPRMT5. Proliferation was analyzed through CCK8 and colony formation assays. Transwell assay was performed to determine cell migration and invasion. Luciferase assay and RIP assay were carried out to analyze the interaction between circPRMT5 and miR-30c.
Results: CircPRMT5 expression was upregulated in PTC tissues and cell lines. And circPRMT5 level was positively linked with advanced stage and lymph node metastasis. CircPRMT5 knockdown inhibited proliferation, migration and invasion while inducing apoptosis. CircPRMT5 worked as a competing endogenous RNA for miR-30c. By inhibiting miR-30c, circPRMT5 promoted the expression of E2F3.
Conclusion: Our findings demonstrate that circPRMT5 acts as an oncogenic circRNA to promote PTC progression via regulating miR-30c/E2F3 axis.
Keywords: circRNA, papillary thyroid cancer, circPRMT5, miR-30c, E2F3
Introduction
Thyroid cancer is a very common malignancy worldwide and its incidence has increased quickly in the past years.1 Papillary thyroid cancer (PTC) is the most common type among thyroid cancer patients and accounts for about 85%.2 Although the prognosis of PTC is usually good and it has a high overall survival, it still impairs the quality of patients because of metastatic potential.3 Notably, the survival rate of PTC patients with advanced stages is under 60%.4 Thus, there is still an urgent requirement to determine novel therapeutic targets for improving the intervention efficiency.
Circular RNA (circRNA) is a new type of noncoding RNAs and characterized by a covalently closed loop.5 circRNA is formed by back-splicing and very stable.6 Increasing evidences show that circRNA has very important functions in tumorigenesis, including PTC.7 Aberrant expression of circRNA may lead to development of several cancers. For example, circRNA_104433 is involved in the regulation of gastric cancer proliferation via inhibiting miR-497-5p.8 CircRNA circHUWE1 knockdown inhibits growth and metastasis of colorectal cancer cells through targeting miR-486.9 circRIP2 promotes bladder cancer cell proliferation, migration and invasion by regulating miR-1305/Tgf-β2/smad3 axis.10 In addition, hsa_circ_001680 regulates growth and metastasis in colorectal cancer cells and affects chemoresistance through miR-340/BMI1 axis.11 However, how circRNA dysregulation affects PTC development remains undefined.
CircPRMT5 was reported to increase invasion of bladder cancer and promote epithelial-mesenchymal transition through targeting miR-30c.12 Besides, circPRMT5 also increases growth of colorectal cancer cells.13 Whether circPRMT5 exerts a role in PTC remains elusive. In this study, we found that circPRMT5 was upregulated in PTC tissues and circPRMT5 knockdown inhibited proliferation and metastasis of PTC cells. We found that circPRMT5 is a competing endogenous RNA (ceRNA) for miR-30c and activates E2F3 expression. Rescue assays demonstrated the roles of circPRMT5/miR-30c/E2F3 axis in PTC. Summarily, our finding reveals the novel function of circPRMT5 in PTC and suggests circPRMT5 may be a potential therapeutic target.
Materials and Methods
Patient Tissue
55 PTC tissues and adjacent normal controls were obtained from Wenling First People’s Hospital. This study was approved by the Ethics Committee of Wenling First People’s Hospital and all experiments were conducted in accordance with the Declaration of Helsinki. All tissues were stored in liquid nitrogen until use. Written informed consents were obtained from patients.
Cell Culture and Transfection
PTC cell lines (K1, TPC-1, IHH4 and BCPAP cells) and the human thyroid normal cell line Nthy-ori 3–1 were from ATCC (Manassas, VA, USA) and cultured using RPMI 1640 (Invitrogen, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen, CA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. siRNAs, miR-30c mimics, miR-30c inhibitors and corresponding negative controls were obtained from GenePharma. Cell transfection was carried out using Lipofectamine 3000 (Invitrogen, CA, USA) at the concentration of 100 nM. After 48 h of transfection, the transfection efficiency was measured by qRT-PCR and cells were then used for experiments.
qRT-PCR
Total RNA was purified using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was reverse-transcribed into cDNA by a Prime Script RT Reagent Kit (Takara, Dalian, China). And qPCR was conducted with SYBR Premix Ex Taq (Takara). U6 or GAPDH was used as an internal control. The expression of circPRMT5 was normalized to GAPDH and calculated according to the 2−ΔΔCt method. The primer sequences were listed: circPRMT5 (5ʹ-CCACTGTACTCCTCTGTGTGT-3ʹ and 5ʹ-CCACTGTACTCCTCTGTGTGT-3ʹ), miR-30c (5ʹ-ACCATGCTGTAGTGTGTGTAAACA-3ʹ and 5ʹ-TCCATGGCAGAAGGAGTAAA-3ʹ), E2F3 (5ʹ-TGACCCAAT GGTAGGCACAT-3ʹ and 5ʹ-CATCTAGGACCACACCGACA-3ʹ) and GAPDH (5ʹ-TGCACCACCAACTGCTTAGC-3ʹ and 5ʹ-GGCATGGACTGTGGTCATGAG-3ʹ).
Cell Viability Assay
Transfected cells were plated into the 96-well plate and cultured for 24h, 48h and 72h, followed by treated with 10μL CCK8 solution for 2h according to the manufacturer’s instructions. Absorbance at 450 nm was determined by a microplate reader (BioTek Instruments, Winooski, VT, USA).
Colony Formation Assay
500 cells were plated into the 6-well plate and cultured using complete medium (10% FBS) for 14 days. Then cells were fixed with 75% ethanol for 1 h, followed by staining with 0.2% crystal violet for 1 h. Colony number was finally counted.
Transwell Assay
Migration and invasion were measured using transwell assay by 8 μm pore transwell chambers either without Matrigel (for migration assays) or with Matrigel (for invasion assays).14 For cell culture, the upper chamber was filled with serum-free medium and the lower chamber was filled with complete medium (10% of FBS).
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using the Magna RIP kit (Millipore, Bedford, MA, USA) according to the manufacturer’s protocol. Briefly, cell lysate was incubated with protein A/G magnetic beads conjugated IgG or Ago2 antibody (Millipore, Billerica, MA, USA) overnight. Then enrichment fold was analyzed by qRT-PCR.
Dual-Luciferase Reporter Assay
Luciferase reporter vectors were constructed as previously described.12 For luciferase reporter assay, reporter vector and miR-30c mimics were co-transfected into cells for 48 h. Then luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, WI, USA) according to the manufacturer’s instructions.
Statistical Analysis
Results were analyzed using PSS ver. 19.0 (SPSS, Chicago, IL, USA) and expressed as s the mean ± SD. One-way ANOVA or Student’s t-test was performed for comparisons. P < 0.05 was considered as statistically significant.
Results
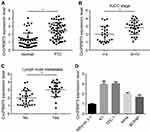
CircPRMT5 Is Upregulated in PTC Tissues
We first analyzed circPRMT5 expression and found that circPRMT5 level was increased in PTC tissues compared with adjacent normal tissues (Figure 1A). Moreover, circPRMT5 expression was positively correlated with AJCC stage and metastasis in PTC tissues (Figure 1B and C). In consistent, we found that circPRMT5 expression was also upregulated in PTC cell lines (Figure 1D).
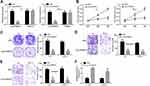
Effects of CircPRMT5 Knockdown on PTC Cells
To analyze the possible function of circPRMT5, we used siRNA to knock down circPRMT5 in K1 and TPC-1 cells (Figure 2A). Notably, circPRMT5 knockdown did not affect the expression of PRMT5 (Figure 2A). Then, CCK8 assay was performed. As shown, circPRMT5 knockdown attenuated the proliferation of K1 and TPC1 cells (Figure 2B). CircPRMT5 knockdown also reduced the colony numbers (Figure 2C). To confirm the correlation between circPRMT5 expression and metastasis, we performed transwell assay. The results showed that circPRMT5 knockdown decreased the migrated and invaded cell numbers (Figure 2D and E). Interestingly, we also observed that circPRMT5 knockdown induced apoptosis (Figure 2F) as the activity of Caspase3 was increased after circPRMT5 depletion. Thus, circPRMT5 knockdown prevents PTC proliferation, migration and invasion.
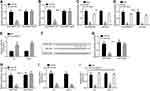
CircPRMT5 Promotes E2F3 Expression via Targeting miR-30c
Previously, circPRMT5 was shown to sponge miR-30c in bladder cancer.12 Through luciferase reporter assay, we also found that miR-30c mimics inhibited the activity of circPRMT5-WT vector (Figure 3A and B), indicating that circPRMT5 also sponges miR-30c in PTC cells. Moreover, through RIP assay, we found that circPRMT5 interacted with miR-30c in K1 and TPC-1 cells (Figure 3C and D). CircPRMT5 knockdown also upregulated miR-30c levels in PTC cells (Figure 3E). We then analyzed the target gene of miR-30c via TargetScan7 tool. We found that E2F3 is the most potential target. We then designed E2F3 luciferase reporter vectors (Figure 3F). We found that miR-30c mimics inhibited the activity of E2F3-WT vector (Figure 3G and H). Furthermore, miR-30c mimics inhibited the expression of E2F3 in K1 and TPC-1 cells (Figure 3I). Notably, circPRMT5 knockdown also inhibited E2F3 expression while miR-30c inhibitors reversed it (Figure 3J), suggesting a circPRMT5/miR-30c/E2F3 regulatory axis in PTC.
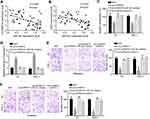
CircPRMT5 Promotes PTC Progression via Targeting miR-30c/E2F3 Axis
We observed that miR-30c expression was negatively correlated with circPRMT5 and E2F3 in PTC tissues (Figure 4A and B). To determine whether circPRMT5 relied on miR-30cE2F3 axis to affect PTC progression, we performed rescue assays. CCK8 assay showed that miR-30c inhibitors or E2F3 overexpression reversed proliferation of circPRMT5-silenced cells (Figure 4C). We also observed similar results of apoptosis, migration and invasion (Figure 4D–F). Therefore, circPRMT5 contributes to PTC development relying on miR-30c/E2F3 signaling.
Discussion
The incidence of PTC is growing gradually during the past decade. Many PTC patients with tumor metastasis show poor prognosis. The mechanism regulating PTC severity remains elusive. CircRNA dysregulation has been demonstrated to participate in cancer development. In our study, we showed that circPRMT5 expression was upregulated in PTC tissues and correlated with metastasis. We found that circPRMT5 downregulation attenuated the growth and metastasis of PTC cells. We also showed that circPRMT5 is a ceRNA for miR-30c to upregulate E2F3 expression.
Growing studies have indicated that circRNA act as a crucial role in cancers, including PTC development.14 For instance, hsa_circ_0039411 initiates PTC development and enhances cancer cell metastasis via miR-1179/ABCA9 axis and miR-1205/MTA1 axis.15 CircRNA circBACH2 upregulation enhances proliferation, migration and invasion of PTC cells through targeting miR-139 to regulate LMO4 level.16 Besides, circ_0025033 is upregulated in PTC tissues and positively correlated with tumor growth and invasion by inhibiting both miR-1231 and miR-1304.17 CircPRMT5 was involved in bladder cancer and colon cancer.12,13 However, whether circPRMT5 regulates PTC needs investigation. In this work, we found that circPRMT5 was upregulated in PTC tissues and cell lines. Moreover, circPRMT5 downregulation suppressed proliferation, migration and invasion of PTC cells. Our results indicate circPRMT5 is an oncogene in PTC. Moreover, we found that circPRMT5 was upregulated in PTC tissues. CircPRMT5 expression levels were positively correlated with AJCC stage and lymph node metastasis, suggesting that circPRMT5 may have potential clinical roles.
CircRNA has been demonstrated to be important sponges for miRNA.18 For example, hsa_circ_0060060 is reported to sponge miR-144-3p to regulate thyroid cancer cisplatin-resistance.19 CircRNA circ-ITCH is found to target miR-22-3p and suppress PTC aggravation.20 CircPRMT5 is also demonstrated to inhibit miR-30c and miR-377.12,13 In our study, we also validated that miR-30c was sponged by circPRMT5. We demonstrated their direct interaction through luciferase reporter assay and RIP assay. Moreover, we showed that circPRMT5 expression was reversely correlated with miR-30c in PTC tissues. MiR-30c has been proven as a key regulator in cancers, such as non-small cell lung cancer, endometrial cancer and hepatocellular carcinoma.21–23 However, how miR-30c affects PTC progression is undefined. We showed that miR-30c inhibitors reversed the effects of circPRMT5 knockdown. Our findings demonstrated that miR-30c is suppressed by circPRMT5 and works as anti-tumor roles.
CircRNA-miRNA-mRNA interactions have been widely validated in tumor development.24 Through TargetScan prediction, we identified that E2F3 is the most potential target of miR-30c. We also confirmed their interaction via luciferase reporter assay. Moreover, we showed that E2F3 expression was inhibited by either circPRMT5 silencing or miR-30c mimics. And miR-30c inhibitors promoted E2F3 expression in circPRMT5-depleted PTC cells. E2F3 is a crucial oncogene in several tumors. For instance, E2F3 promotes proliferation, migration and invasion in nasopharyngeal cancer.25 E2F3 collaborates with NF-κB pathway to aggravate bladder cancer.26 Additionally, E2F3 also promotes ovarian cancer growth and chemo-sensitivity.27 Nevertheless, E2F3 function in PTC is not clear. In our study, we found that E2F3 overexpression restored the abilities of proliferation, migration and invasion in circPRMT5-depleted cells. Thus, E2F3 is an oncogene in PTC.
Conclusively, our findings showed that circPRMT5 promotes PTC progression through targeting miR-30c/E2F3 pathway. And circPRMT5/miR-30c/E2F3 axis may be a novel therapeutic target for PTC intervention.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lubitz CC, Sosa JA. The changing landscape of papillary thyroid cancer: epidemiology, management, and the implications for patients. Cancer. 2016;122(24):3754–3759. doi:10.1002/cncr.30201
2. Vitale G, Caraglia M, Ciccarelli A, et al. Current approaches and perspectives in the therapy of medullary thyroid carcinoma. Cancer. 2001;91(9):1797–1808. doi:10.1002/1097-0142(20010501)91:9<1797::AID-CNCR1199>3.0.CO;2-P
3. Bongiovanni M, Sykiotis GP, La Rosa S, et al. Macrofollicular variant of follicular thyroid carcinoma: a rare underappreciated pitfall in the diagnosis of thyroid carcinoma. Thyroid. 2020;30(1):72–80. doi:10.1089/thy.2018.0607
4. Owonikoko TK, Hossain MS, Bhimani C, et al. Soluble FAS ligand as a biomarker of disease recurrence in differentiated thyroid cancer. Cancer. 2013;119(8):1503–1511. doi:10.1002/cncr.27937
5. Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18(1):303. doi:10.1186/s12885-018-4213-0
6. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
7. Lan X, Xu J, Chen C, et al. The landscape of circular RNA expression profiles in papillary thyroid carcinoma based on RNA sequencing. Cell Physiol Biochem. 2018;47(3):1122–1132. doi:10.1159/000490188
8. Wei W, Mo X, Yan L, et al. Circular RNA profiling reveals that circRNA_104433 regulates cell growth by targeting miR-497-5p in gastric cancer. Cancer Manag Res. 2020;12:15–30. doi:10.2147/CMAR.S219307
9. Chen H-Y, Li X-N, Ye C-X, Chen Z-L, Wang Z-J. Circular RNA circHUWE1 is upregulated and promotes cell proliferation, migration and invasion in colorectal cancer by sponging miR-486. Onco Targets Ther. 2020;13:423–434. doi:10.2147/OTT.S233338
10. Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-beta2/smad3 pathway. Mol Cancer. 2020;19(1):23. doi:10.1186/s12943-019-1129-5
11. Jian X, He H, Zhu J, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19(1):20. doi:10.1186/s12943-020-1134-8
12. Chen X, Chen RX, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–6330. doi:10.1158/1078-0432.CCR-18-1270
13. Yang B, Du K, Yang C, et al. CircPRMT5 circular RNA promotes proliferation of colorectal cancer through sponging miR-377 to induce E2F3 expression. J Cell Mol Med. 2020;24(6):3431–3437. doi:10.1111/jcmm.15019
14. Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 2019;38(1):318. doi:10.1186/s13046-019-1321-x
15. Yang Y, Ding L, Li Y, Xuan C. Hsa_circ_0039411 promotes tumorigenesis and progression of papillary thyroid cancer by miR-1179/ABCA9 and miR-1205/MTA1 signaling pathways. J Cell Physiol. 2020;235(2):1321–1329. doi:10.1002/jcp.29048
16. Cai X, Zhao Z, Dong J, et al. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10(3):184. doi:10.1038/s41419-019-1439-y
17. Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun. 2019;510(2):334–338. doi:10.1016/j.bbrc.2019.01.108
18. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1):27. doi:10.1186/s12943-019-0951-0
19. Liu F, Zhang J, Qin L, et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging (Albany NY). 2018;10(12):3806–3820. doi:10.18632/aging.101674
20. Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/beta-catenin pathway. Biochem Biophys Res Commun. 2018;504(1):283–288. doi:10.1016/j.bbrc.2018.08.175
21. Zhou Y, Shi H, Du Y, et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging (Albany NY). 2019;11(18):7386–7401. doi:10.18632/aging.102226
22. Xu X, Kong X, Liu T, et al. Metastasis-associated protein 1, modulated by miR-30c, promotes endometrial cancer progression through AKT/mTOR/4E-BP1 pathway. Gynecol Oncol. 2019;154(1):207–217. doi:10.1016/j.ygyno.2019.04.005
23. Zhang J, Cai M, Jiang D, Xu L. Upregulated LncRNA-CCAT1 promotes hepatocellular carcinoma progression by functioning as miR-30c-2-3p sponge. Cell Biochem Funct. 2019;37(2):84–92. doi:10.1002/cbf.3375
24. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4. doi:10.1186/s12943-018-0933-7
25. Wang T, Du M, Zhang W, et al. MicroRNA-432 suppresses invasion and migration via E2F3 in nasopharyngeal carcinoma. Onco Targets Ther. 2019;12:11271–11280. doi:10.2147/OTT.S233435
26. Shi F, Deng Z, Zhou Z, et al. QKI-6 inhibits bladder cancer malignant behaviours through down-regulating E2F3 and NF-kappaB signalling. J Cell Mol Med. 2019;23(10):6578–6594. doi:10.1111/jcmm.14481
27. Jin Y, Wei J, Xu S, Guan F, Yin L, Zhu H. miR2103p regulates cell growth and affects cisplatin sensitivity in human ovarian cancer cells via targeting E2F3. Mol Med Rep. 2019;19(6):4946–4954. doi:10.3892/mmr.2019.10129
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.