Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA circNELL2 Acts as the Sponge of miR-127-5p to Promote Esophageal Squamous Cell Carcinoma Progression
Authors Xiong G, Diao D, Lu D, Liu X, Liu Z, Mai S, Feng S, Dong X, Cai K
Received 31 January 2020
Accepted for publication 6 June 2020
Published 16 September 2020 Volume 2020:13 Pages 9245—9255
DOI https://doi.org/10.2147/OTT.S247847
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Gang Xiong, Dingwei Diao, Di Lu, Xiguang Liu, Zhaoguo Liu, Shijie Mai, Siyang Feng, Xiaoying Dong, Kaican Cai
Department of Thoracic Surgery, Southern Medical University Nanfang Hospital, Guangzhou City, Guangdong Province 510515, People’s Republic of China
Correspondence: Kaican Cai
Department of Thoracic Surgery, Southern Medical University Nanfang Hospital, No. 1838, Guangzhoudadao Road, Guangzhou City, Guangdong Province 510515, People’s Republic of China
Email [email protected]
Introduction: Owing to its involvement in both the initiation and progression of various cancers, aberrant circular RNA (circRNA) expression has been researched extensively in the recent times. In the present study, we aim to investigate the effect of a novel circRNA has_circ_0025933 (circNELL2) in the progression of esophageal squamous cell carcinoma (ESCC).
Materials and Methods: Sanger sequencing and the detection of circNELL2 level after RNase R or actinomycin D treatment were performed to identify the existence of cirNELL2 in ESCC cells. WST, EDU staining and colony-formation assay were used to assess the proliferation while transwell assay was used to evaluate the migration of ESCC cells. Luciferase assay, RNA pull down and the FISH assay were performed to verify the interaction between circNELL2 and miR-127-5p as well as miR-127-5p and CDC6. Xenograft model was carried out to evaluate the effect of circNELL2 in vivo.
Results: circNELL2 was proved to exist in ESCC cells. The up-regulated expression of circNELL2 in the clinical ESCC specimens was also verified. Next, function studies suggested that circNELL2 knockdown inhibited the proliferation of ESCC cells in vitro and in vivo, while circNELL2 overexpression promotes that of ESCC cells. Besides, this study mechanically predicted and verified the target miR of circNELL2, which is miR-127-5p. It was found that miR-127-5p was capable of reversing the effect of circNELL2 on ESCC cells. Moreover, miR-127-5p was also found to target CDC6 to participate in the regulation of cell phenotype.
Discussion: circNELL2 promoted the progression of ESCC cells via sponging miR-127-5p, and it has the potential to serve as a novel prognostic and therapeutic target for ESCC.
Keywords: circular RNA circNELL2, miR-127-5p, esophageal squamous cell carcinoma
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most common esophageal carcinoma accounts for more than 90% of it, with its incidence rising continuously in the recent years. Conventional therapeutic strategies of ESCC include surgery, hormone therapy and radioactive iodine therapy.1,2 Despite the advance in the development of these diagnosis and treatment strategies, the 5-year survival rate of patients with advanced ESCC is still under 60%.3 Thus, it is urgent to discover novel potential therapeutic targets which require us to have a much more comprehensive understanding of the molecular mechanism underlying ESCC progression.
Circular RNAs (circRNA) are a novel family of non-coding RNAs characterized by formation of closed loops without 5ʹ to 3ʹ polarity or polyadenylated tails covalently.4 They are highly conserved and abundant through different species for their stable structure which can resist the digestion of RNase R making them idea biomarkers for diagnosis and prognosis.5,6 Since early discovered, circRNAs were identified as splicing errors or by-product of incorrect RNA splicing with no value. However, with the deepening of research on them, they have been demonstrated to play crucial roles in the biological processes such as splicing regulation,7 sponging microRNA (miRNA),8,9 and regulating protein–RNA interactions.10 As in the field of cancer, circRNAs have been indicated to be dysregulated in various cancers including prostate cancer,11 colorectal cancer,12 hepatocellular carcinoma,13,14 as well as glioblastoma,15 and participate in the physiological processes of these cancers. Previous studies have introduced the function of hsa_circ_0000654 and hsa_circ_0000337 in the progression of ESCC through diverse molecular mechanisms.16–18
Here, we identified circNELL2 (hsa_circ_0025933) which was spliced from exon 14 and exon 15 of the neural EGFL like 2 (NELL2) gene, and with the length of 294 nucleotides. In our study, we found the dysregulated expression of circNELL2 and its oncogenic role in ESCC through sponging miR-127-5p and further regulating the downstream CDC6 expression. These findings give us a better understanding of the function of circRNAs in ESCC progression and provide a novel potential therapeutic and diagnosis biomarker.
Materials and Methods
Patient Specimens
Twenty-five pairs of ESCC tissues and adjacent normal tissues were obtained from ESCC patients at the Nanfang Hospital. Upon collection, all samples were snap-frozen using liquid nitrogen and stored at a temperature of −80°C until. All procedures were performed in accordance with the Declaration of Helsinki of the World Medical Association. The written informed consents have been provided by the patients. All experiments involving animals and human tissues were approved by the Ethics Committee of the Nanfang Hospital.
Cell Culture
ESCC cell lines (OE33 and FLO-1) were obtained from the ATCC (Manassas, VA, USA). All obtained cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, CA, USA) containing 10% FBS (fetal bovine serum, Gibco, CA, USA) supplemented with 100 U·mL−1 penicillin, and 100 mg·mL−1 streptomycin (Invitrogen, CA, USA) in humid conditions with 5% CO2 at a temperature of 37°C.
Plasmids Construction
SiRNAs for circNELL2, the control, the miR-127-5p mimic, mimic control, miR-127-5p inhibitor, and inhibitor control are designed and synthesized by GenePharma (Shanghai, China). The shRNA against circNELL2 and negative control were also synthesized by GenePharma (Shanghai, China). In order to establish the over-expressing vectors of circNELL2 and CDC6, the coding sequence of human circNELL2 and CDC6 were synthesized and inserted into the pLCDH-ciR and pcDNA3 vector, respectively (Bioco Biotech Co, Tianjin, China). Furthermore, wild type and mutant sequence that contains the binding site of miR-127-5p from circNELL2 and CDC6 were synthesized and cloned into pGL3 luciferase reporter vectors (Promega, CA, USA).
Oligonucleotides Transfection and Stable Transfection
Lipofectamine 3000 (Invitrogen, CA, USA) was used to transfect Oligonucleotides at a concentration of 60 nmol/L, the procedure was done in accordance with the manufacturer’s instructions. In order to establish stable cell lines, all the cells subjected to transfection were subsequently selected with 2 μg/mL puromycin continuously for a period of 2 weeks.
WST Assay
WST assay was performed to evaluate the cell viability, the procedure followed instructions from the manufacturer. The cells were seeded in 96-well plates (5×103/well). Following the treatment, cell viability was determined every 24 hours over a period of 3 days. Using a Microplate Reader (Bio-Rad, CA, USA), absorbance was detected at 490 nm. Each assay was performed at least three times.
Colony-Formation Assay
The cells were treated with indicated conditions, and then seeded in 12-well plates (100/well). Following an incubation period of 2 weeks, crystal violet (0.05%, Beyotime, Shanghai, China) was used to stain the colonies. Colonies containing more than 50 cells were counted.
Transwell Assay
Transwell assay was used to detect cell invasion ability (Millipore, MA, USA). Cells were treated with indicated conditions, and then the treated cells were seeded on the upper insert coated with 2% Matrigel (BD Biosciences, NY, USA) in 24-well plates. The upper insert was filled with medium lacking serum, and the lower chamber was filled with 600 μL of DMEM supplemented with 10% FBS. Following a period of 24 hrs of incubation, cells invaded to the lower chambers were fixed with methanol, stained with crystal violet.
Luciferase Activity Assay
The wild-type or mutant seed sequence at the predicted region of circNELL2 and CDC6 were synthesized and cloned into the pGL3 Luciferase Reporter vectors (Promega, CA, USA) at the KpnI and BamHI sites. Following this, the ESCC cells were co-transfected with miR-127-5p mimics, or mimic control, together with pGL3 vectors, which contained the WT or mut binding region of circNELL2 and CDC6. In addition, TRL-SV40 plasmid (Promega, CA, USA) was also transfected as a normalizing control. Dual-Luciferase Assay was used to harvest the cells for detecting the activity of luciferase (Promega, WI, USA) at 48 hrs following the transfection.
Quantitative Real-Time PCR
The miRNeasy Mini Kit (Qiagen, Germany) was used to extract mRNA and miRNA from the tissues and cells. cDNA was attained by the means of reverse transcription assay with the use of the reverse transcriptase, and Oligod (T) was employed as the primer. With regards to the cDNA synthesis of miR-127-5p, a particular reverse transcription primer was used. QRT-PCR was carried out using a SYBR-Green Supermix kit in a biorad IQ5 system (CA, USA). U6 small nuclear RNA (U6-snRNA) and GAPDH were used as the internal control for the purpose of evaluating the corresponding relative expression levels of miR-127-5p, circNELL2, and mRNAs.
Western Blotting
RIPA buffer (Jiancheng, Nanjing, China) was used to extract the proteins of tissue samples and cells. The protein extracts were separated on a sodium dodecyl sulfatepolyacrylamide gels (SDS-PAGE), which were further transferred to PVDF membranes. Next, the blots were incubated in 5% nonfat dry milk in phosphate-buffered saline (PBS) with 0.1% Tween 20 for a duration of 2 hrs followed by incubating with primary antibodies (Proteintech, Wuhan, China) overnight at a temperature of 4°C. Next, the blots were incubated in horseradish peroxidase conjugated secondary antibody (Cell Signaling Technology, USA) for a duration of 2 hrs. Following this, the proteins were visualized using an ECL kit (Jiancheng, Nanjing, China), and quantified using the Image J software.
Fluorescence in situ Hybridization (FISH)
Alexa Fluor 555-labeled circNELL2 probes, and Alexa Fluor 488-labeled miR-127-5p probes were designed and synthesized by RiboBio (Guangzhou, China). A fluorescent in situ Hybridization Kit (RiboBio, Guangzhou, China) was used to carry out the FISH experiment. A total of 1×105 cells were seeded onto autoclaved glass slides and cultured for a period of 24 hours. Following a fixing procedure with 4% paraformaldehyde for a duration of 20 minutes, was followed by permeabilization with 0.5% Triton X-100 for a duration of 10 minutes, the cells were then cultured overnight at a temperature of 37°C. Finally, the slides were incubated with DAPI to stain cell nuclear before being observed under a fluorescence microscope (Leica, Wetzlar, Germany).
RNA Pull Down
Biotin-labeled miR-127-5p probe and control probe were synthesized by Sangon Biotech (Shanghai, China). Probe-coated beads were generated by co-incubating the probe with streptavidin-coated beads (Invitrogen, CA, USA) at a temperature of 25°C for 2 hours. ESCC cells were collected and lysed and incubated with miR-127-5p probes overnight at 4°C. Following this, the beads were eluted and the complex was purified with TRIzol (Takara, Dalian, China). Then the abundance of circNELL2 and CDC6 was analyzed using qRT-PCR.
Immunohistochemistry (IHC)
The tumor tissues were fixed in 4% paraformaldehyde for 24 h; next, they were dehydrated in a graded alcohol series and embedded in paraffin. Following this, they were cut into 5μm sections. The sections were deparaffinized, rehydrated with a graded alcohol series, and then incubated in 96°C 0.01 mol/L sodium citrate buffer to allow the retrieval of antigen. Following incubation, 5% H2O2 was applied for quenching endogenous peroxidase activity. In order to reduce non-specific binding, the sections were blocked with 10% non-immune goat serum. The sections were incubated with primary antibodies overnight at 4°C. By following the manufacturer’s instructions (Beyotime, Shanghai, China), immunostaining was performed using streptavidin-peroxidase and diaminobenzidine (DAB). The sections were mounted with gummi after staining the nucleus with hematoxylin, after being dehydrated in a graded alcohol series and pellucidum in xylene.
Xenograft Model
As mentioned above, stably expressing or knocking down circNELL2 cell lines OE-33 and FLO-1 were established. Cells were harvested and resuspended in DMEM medium. The nude mice were injected at the posterior flank with a total of 3 × 106/100 μL cell subcutaneously. Every 3 days, the tumor size was monitored by measuring the length (L) and width (W) with calipers. After 28 days, the tumors were excised from the sacrificed mice and weighed. All experiments involving animals were approved by the Ethics Committee of the Nanfang Hospital and followed by Guidelines for the ethical review of laboratory animal welfare People’s Republic of China National Standard GB/T 35892–2018.
Statistical analysis
All data are presented as the means±standard deviation (SD). The statistical analyses were performed using SPSS 20 software (Abbott Laboratories, Chicago, IL, USA). Data were analyzed with one-way ANOVA and Student’s t test. P<0.05 was considered to be statistically significant. Linear correlation analyses were performed to determine correlations between circNELL2, miR-127-5p and CDC6 expression levels. All experiments were performed independently triplicates.
Results
Identification of circNELL2 in ESCC
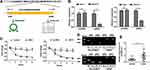
The 294-bp circNELL2 was looped by circularization of exon 14 and 15 of the NELL2 gene, which was located at chromosome 12p22 referring to the sequence from circBase. In the present study, it was further verified by the head-to-tail splicing through sanger sequencing (Figure 1A). It is known that circRNAs are stable for their ring structure. In order to evaluate the stability of circNELL2, the present study evaluated the amount of circNELL2 of OE-33 and FLO-1 cells under RNase R treatment. As indicated in Figure 1B, the amount of linear mRNA of NELL2 was significantly decreased under the RNase R treatment. However, there was no evidence of alteration of circNELL2 under digestion of RNase R. Furthermore, we assessed the levels of linear NELL2 and circNELL2 under the treatment of Actinomycin D. The results revealed that treatment with actinomycin D had no significant effect on the level of circNELL2 at different times; however, the level of linear NELL2 was significantly decreased after actinomycin D (Figure 1C). Furthermore, cDNA and gDNA were extracted separately from OE-33 and FLO-1 cells, and they were subjected to nucleic acid electrophoresis detection. According to the results, circNELL2 could be detected in cDNA; however, they were undetectable in gDNA (Figure 1D). Once it was identified that circNELL2 existed, the next step involved evaluating the expression level of circNELL2 in ESCC. It was significantly up-regulated in the ESCC tissues compared to that in normal tissues (Figure 1E).
Knock Down of circNELL2 Inhibited the Proliferation, Colony Formation, and the Migration in ESCC Cells
Following this, the study carried out loss of function studies including cell proliferation, colony formation, and cell migration. In order to further detect biological functions of circNELL2, two siRNAs targeting the back-splice sequence of circNELL2 was designed to knock down the expression of circNELL2. Firstly, the level of circNELL2 was detected to confirm whether the two siRNAs were capable of inhibiting the expression of circNELL2. According to the results, both siRNAs successfully decreased the expression level of circSMAD4A (Figure 2A). Subsequently, CCK-8 and EDU staining were performed to evaluate the proliferation of OE-33 and FLO-1 cells. A notable inhibition of OE-33 and FLO-1 cell proliferation subjected to circNELL2 knock down was observed (Figure 2B and C). In addition, colony formation and transwell assay indicated that circNELL2 knock down significantly reduced colony-formation ability and cell migration of ESCC cells (Figure 2D and E).
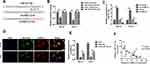
circNELL2 Works as an Efficient miR-127-5p Sponge in ESCC
Generally, it has been established that circRNAs are capable of sponging miRNAs and subsequently abrogate the function of the corresponding miRNAs. On this basis, it was decided to explore whether circNELL2 sponge miRNAs. Circinteractom databases were used to predict the miRNA targets of circNELL2, among which, miR-127-5p was selected as a potential target. Figure 3A shows the target region between circNELL2 and miR-127-5p. Luciferase activity assay in both OE-33 and FLO-1 cells was carried out. The findings indicate that miR-127-5p evidently down-regulated the luciferase activity co-transfected with pGL3-3ʹUTR of circNELL2; however, it does not include the pGL3-3ʹUTR-mut (Figure 3B). qPCR It was further confirmed through analyses that circNELL2 overexpression decreased the level of miR-127-5p, whilst the knock down of circNELL2 increased that of miR-127-5p (Figure 3C). Moreover, the FISH experiment was performed to detect the location of circNELL2 and miR-127-5p. As illustrated in Figure 3D, co-location of circNELL2 and miR-127-5p in cytoplasm was observed. In order to further detect whether circNELL2 could bind directly to miR-127-5p endogenously, RNA pull down was carried. It was found that biotin-labeled miR-127-5p enriched circNELL2 which indicated the binding between circ NELL2 and miR-127-5p (Figure 3E). Pearson analysis indicated a negative correlation between circNELL2 and miR-127-5p (Figure 3F).
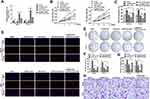
miR-127-5p Reversed the Function of circNELL2 in ESCC
With the aim of investigating the role of miR-127-5p in ESCC, and the relation between miR-127-5p and circNELL2, a rescue experiment was carried out. OE-33 and FLO-1 cells were transfected with circNELL2, miR-127-5p or with both. Firstly, the expression level of miR-127-5p under different kinds of oligonucleotide transfection was evaluated. It was observed that circNELL2 overexpressing vector remarkably decreased the level of miR-127-5p whilst miR-127-5p overexpressing vector increased the level of that. It was also found that the co-transfection of circNELL2 and miR-127-5p reversed the down-regulation of miR-127-5p which was induced by circNELL2 overexpression (Figure 4A). Function studies indicated that circNELL2 had the capability of promoting cell proliferation, colony formation, and migration of ESCC cells. Contrarily, miR-127-5p overexpression inhibited cell proliferation, colony formation, and migration of ESCC cells. Finally, co-transfection of circNELL2 and miR-127-5p reversed the effect of circNELL2 on cell proliferation, colony formation, and migration of ESCC cells (Figure 4B-H).
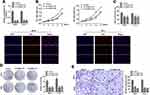
miR-127-5p Directly Targets CDC6 in ESCC
In order to predict the targets of miR-127-5p, Miranda and TargetScan databases were used. Among these, CDC6 was selected as a potential target due to its existence in both databases (Figure 5A). Figure 5B shows the target region between miR-127-5p and CDC6. Luciferase activity assay was carried out in both OE-33 and FLO-1 cells. As indicated by the findings, miR-127-5p evidently down-regulated the luciferase activity co-transfected with pGL3-3ʹUTR of CDC6; however, the pGL3-3ʹUTR-mut was not found to be so (Figure 5C). The qPCR and Western blot analyses further confirmed that miR-127-5p overexpression decreased the level of CDC6, it was also revealed that knock down of miR-127-5p increased the level of CDC6 (Figure 5D, E). In order to further detect whether miR-127-5p could directly bind to CDC6 endogenously, RNA pull down was carried again. It was also found that biotin-labeled miR-127-5p enriched CDC6 which indicated the binding between CDC6 and miR-127-5p (Figure 5F). qPCR results revealed that circNELL2 silencing down-regulated the expression of CDC6 (Figure 5G). Pearson analysis showed a negative correlation between miR-127-5p and CDC6, and a positive correlation between circNELL2 and CDC6 (Figure 5H and I).
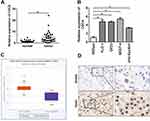
CDC6 Was Up-Regulated in ESCC
In order to further explore the potential effect of CDC6 in ESCC, the expression level of CDC6 in ESCC tissues was evaluated. Firstly, qPCR analysis was carried out, and it was found that CDC6 was significantly up-regulated in ESCC tissues compared to that in normal tissues (Figure 6A) and cell lines (Figure 6B). Additionally, pan-cancer analysis was performed using the data from the StarBase dataset, the analysis showed a notable elevation of CDC6 level in ESCC specimens (Figure 6C). Further confirmation of the up-regulated expression of CDC6 in ESCC was provided by IHC analysis (Figure 6D).
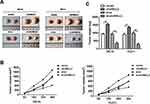
circNELL2 Affects ESCC Growth and Proliferation in vivo
In order to explain the role of circNELL2 in the regulation of ESCC, the xenograft nude mice model was developed by means of subcutaneously injecting genetically modified OE-33 and FLO-1 cells. When compared with the vector and si-nc group, the circNELL2 overexpressing vector transfection substantially promoted the tumor growth of ESCC, this is evident from both the tumor growth curve and the weight of the tumor (Figure 7A–C).
Discussion
Circular RNAs are a novel class of non-coding RNAs, they have not been extensively studied so far in ESCC. In the present study, a novel circular RNA named circNELL2 base was identified. It was known to up-regulate in human ESCC tissues and cell lines.19 Functionally, this study demonstrated that knockdown of circNELL2 has the potential to inhibit cell proliferation as well as migration in vitro and in vivo, contrastingly, the up-regulation of circNELL2 exerted the opposite effects. Mechanistically, circNELL2 could function as a ceRNA through harboring miR-127-5p to abolish the suppressive effect on the target gene CDC6 in ESCC progression.
Circular RNAs participate in the regulation of physiological or physiological processes of cancer cells through its main mechanism of acting as a sponge for microRNAs.8 In the previous studies, circRNAs such as hsa_circ_0000654, hsa_circ_0000337, and circ-PRKCI have been reported to possess biological functionalities in ESCC by binding to miR-149-5p, miR-670-5p, and miR-3680-3p correspondingly. It was first identified that the role of miR-127-5p in ESCC was a target miRNA of circNELL2.
There are relatively few studies on miR-127-5p. It can be sponged by other circRNAs such as circ_0136474,20 circRNA.33186,21 and CircUSP45.22 The research of miR-127-5p is mainly in the field of osteology. MiR-127-5p contributes to the pathogenesis of Osteoarthritis via targeting circRNA.33186 and CircUSP45.21–23 In addition, it regulates osteopontin (OPN)-mediated proliferation of human chondrocytes through PI3K/Akt pathway.24 For example, miR-127 was found to be downregulated in breast cancer.25 MicroRNA-127-5p targets the biliverdin reductase B/nuclear factor-κB pathway to suppress cell growth in hepatocellular carcinoma cells.26
In the present study, it was firstly found that miR-127-5p was down-regulated in ESCC tissues and cell lines, it was also found to be negatively correlated with circNELL2. Gain of function study showed an anti-cancer effect of miR-127-5p in ESCC which has not been elucidated before. In order to further investigate the effect of miR-127-5p in ESCC, a loss of function and in vivo study should be carried out following this research. In addition, CDC6 was predicted and verified as the direct target of miR-127-5p.
Cell division cycle 6 (Cdc6) is a vital mediator of DNA replication, it plays a crucial role in the activation and maintenance of the cell cycle.27 The best-characterized function of Cdc6 is the assembly of pre-replicative complexes (pre-RC) at origins to initiate DNA replication in the G1 phase.28–30 Even though Cdc6 is widely known to be correlated with the progression of human cancers,8,17-19 the exact effect and mechanism of Cdc6 on the development of cancer is yet to be elucidated. Particularly, the role of CDC6 has not been studied before.
It was demonstrated that CDC6 was significantly up-regulated due to the qPCR and IHC results in addition to the data analysis based on Star Base. The present study speculates that CDC6 exerts a critical role in the development of ESCC. However, further functional studies should be carried out to conclusively prove it. Furthermore, it was also confirmed that miR-127-5p targets CDC6 in ESCC which is a unique outcome of this study. Rescue experiments should also be performed to verify this relationship.
In summary, the current investigation shed light on the critical role of circNELL2 and miR-127-5p in the progression of ESCC by the means of regulating cell proliferation, colony formation, and migration. There have not been any prior studies that have thoroughly investigated the biological functions of circNELL2, and the underlying mechanism in ESCC as performed in this study. The findings in this study are likely to provide potential novel targets such as circNELL2 or miR-127-5p for effective therapies against esophageal squamous cell carcinoma.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Agrawal N, Akbani R, Aksoy B. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi:10.1016/j.cell.2014.09.050
2. Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–199. doi:10.1038/nrc3431
3. Wu DM, Wang S, Wen X, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9(10):947. doi:10.1038/s41419-018-0975-1
4. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
5. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi:10.1080/15476286.2016.1220473
6. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
7. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
8. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
9. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi:10.1093/nar/gkw027
10. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
11. Chen S, Huang V, Xu X, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4):831–843. doi:10.1016/j.cell.2019.01.025
12. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi:10.1158/0008-5472.CAN-16-1883
13. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi:10.1016/j.jhep.2018.01.012
14. Han J, Meng J, Chen S, et al. YY1 complex promotes quaking expression via super-enhancer binding during EMT of hepatocellular carcinoma. Cancer Res. 2019;79(7):1451–1464. doi:10.1158/0008-5472.CAN-18-2238
15. Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78(17):4812–4825. doi:10.1158/0008-5472.CAN-18-0532
16. Xu Z, Tie X, Li N, Yi Z, Shen F, Zhang Y. Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. IUBMB Life. 2019.
17. Song H, Xu D, Shi P, et al. Upregulated circ RNA hsa_circ_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma. Cancer Manag Res. 2019;11:1997–2006.
18. Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-kappaB signals. Cancer Biol Ther. 2019;20(1):73–80. doi:10.1080/15384047.2018.1507254
19. Song J, Lu Y, Sun W, Han M, Zhang Y, Zhang J. Changing expression profiles of lncRNAs, circRNAs and mRNAs in esophageal squamous carcinoma. Oncol Lett. 2019;18(5):5363–5373. doi:10.3892/ol.2019.10880
20. Li Z, Yuan B, Pei Z, et al. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J Cell Mol Med. 2019;23(10):6554–6564. doi:10.1111/jcmm.14400
21. Zhou ZB, Huang GX, Fu Q, et al. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 2019;27(3):531–541. doi:10.1016/j.ymthe.2019.01.006
22. Kuang MJ, Xing F, Wang D, Sun L, Ma JX, Ma XL. CircUSP45 inhibited osteogenesis in glucocorticoid-induced osteonecrosis of femoral head by sponging miR-127-5p through PTEN/AKT signal pathway: experimental studies. Biochem Biophys Res Commun. 2019;509(1):255–261. doi:10.1016/j.bbrc.2018.12.116
23. Jakob F, Tony HP, Schneider D, Thole HH. Immunological detection of the oestradiol receptor protein in cell lines derived from the lymphatic system and the haematopoietic system: variability of specific hormone binding in vitro. J Endocrinol. 1992;134(3):397–404. doi:10.1677/joe.0.1340397
24. Liang J, Xu L, Zhou F, et al. MALAT1/miR-127-5p regulates osteopontin (OPN)-mediated proliferation of human chondrocytes through PI3K/Akt pathway. J Cell Biochem. 2018;119(1):431–439. doi:10.1002/jcb.26200
25. Pronina IV, Loginov VI, Burdennyy AM, et al. DNA methylation contributes to deregulation of 12 cancer-associated microRNAs and breast cancer progression. Gene. 2017;604:1–8. doi:10.1016/j.gene.2016.12.018
26. Huan L, Bao C, Chen D, et al. MicroRNA-127-5p targets the biliverdin reductase B/nuclear factor-kappaB pathway to suppress cell growth in hepatocellular carcinoma cells. Cancer Sci. 2016;107(3):258–266. doi:10.1111/cas.12869
27. Borlado LR, Mendez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis. 2008;29(2):237–243. doi:10.1093/carcin/bgm268
28. Murphy N, Ring M, Heffron CC, et al. Quantitation of CDC6 and MCM5 mRNA in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Mod Pathol. 2005;18(6):844–849. doi:10.1038/modpathol.3800361
29. Karakaidos P, Taraviras S, Vassiliou LV, et al. Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: synergistic effect with mutant p53 on tumor growth and chromosomal instability–evidence of E2F-1 transcriptional control over hCdt1. Am J Pathol. 2004;165(4):1351–1365. doi:10.1016/S0002-9440(10)63393-7
30. Feng CJ, Li HJ, Li JN, Lu YJ, Liao GQ. Expression of Mcm7 and Cdc6 in oral squamous cell carcinoma and precancerous lesions. Anticancer Res. 2008;28(6A):3763–3769.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.