Back to Journals » Cancer Management and Research » Volume 13
Circular RNA circ_0020123 Promotes Non-Small Cell Lung Cancer Progression Through miR-384/TRIM44 Axis
Authors Ma Q, Huai B, Liu Y, Jia Z, Zhao Q
Received 28 August 2020
Accepted for publication 20 November 2020
Published 7 January 2021 Volume 2021:13 Pages 75—87
DOI https://doi.org/10.2147/CMAR.S278913
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Qingshan Ma,1 Baogang Huai,2 Yuting Liu,3 Zhongyao Jia,1 Qilong Zhao1
1Department of Oncology, Linyi People’s Hospital, Linyi, Shandong 276000, People’s Republic of China; 2Department of Pulmonary Disease, Pinyi County Hospital of Traditional Chinese Medicine, Linyi, Shandong 273300, People’s Republic of China; 3University Department, Linyi People’s Hospital, Linyi, Shandong 276000, People’s Republic of China
Correspondence: Qilong Zhao
Department of Oncology, Linyi People’s Hospital, No. 27, Jiefang Road, Linyi, Shandong 276000, People’s Republic of China
Tel +86-539 8078518
Email [email protected]
Background: It was reported that circular RNAs (circRNAs) and microRNAs (miRNAs) were related to non-small cell lung cancer (NSCLC) development. However, the detailed mechanisms of circ_0020123 and miR-384 in NSCLC are elusive.
Methods: QRT-PCR and Western blot assay were performed to detect the transcription and protein levels of genes, respectively. Then, the functional experiments, including MTT assay, flow cytometry, and transwell assay, were employed. Besides, the interaction between miR-384 and circ_0020123 or tripartite motif‑containing protein 44 (TRIM44) was predicted by starbase or targetscan, and then verified by the dual-luciferase reporter, RNA pull-down assays and RNA immunoprecipitation assay (RIP). Mouse xenograft assay was performed to evaluate the effect of circ_0020123 on tumor growth in vivo.
Results: Levels of circ_0020123 and TRIM44 were enhanced, and the miR-384 level was attenuated in NSCLC tissues and cells. Circ_0020123 depletion attenuated the abilities of NSCLC cell viability, migration, invasion, and epithelial–mesenchymal transition (EMT), and induced apoptosis. Besides, circ_0020123 interacted with miR-384, and miR-384 targeted TRIM44. Circ_0020123 regulated cell progression by regulating miR-384 and subsequently mediated TRIM44 expression. Besides, circ_0020123 depletion repressed tumor growth in vivo.
Conclusion: We demonstrated that circ_0020123 knockdown suppressed NSCLC cell progression by regulating the miR-384/TRIM44 axis, providing the theoretical basis for the therapy of NSCLC.
Keywords: circ_0020123, miR-384, TRIM44, cell growth, NSCLC
Introduction
Non-small cell lung cancer (NSCLC) was demonstrated to contribute to nearly 1.4 million deaths every year.1 Over the past decades, treatment options made great progress, but the NSCLC 5-year survival rate is still low.2 Moreover, the recurrence rate for NSCLC is high, according to the statistics in 2018.3 Therefore, investigating the mechanism of NSCLC development is necessary for the therapy of NSCLC patients.
Circular RNAs (circRNAs), a novel group of conserved RNAs, exert pivotal functions in mammalian cells.4 Present evidence reveals that circRNAs regulate miRNA levels as the sponges for miRNAs or alter gene transcription by binding to the RNA-binding proteins.5,6 In recent years, circRNAs are reported to be closely related to many diseases, including cancer,7 diabetes,8 and cardiovascular disease.9 CircRNAs are essential for the development of NSCLC. For example, enhanced circ_0020123 was identified as a positive regulator for NSCLC development.10 However, the underlying mechanism of circ_0020123 in NSCLC remains unclear.
With approximately 22 nucleotides, microRNAs (miRNAs) could regulate downstream gene expression.11 Numerous miRNAs were observed as the tumor suppressor and were low expressed in NSCLC.12–14 For instance, miR-384 inhibited cell development and mobility through regulating the wingless/integrated (Wnt) pathway in NSCLC cells.12 Guo et al confirmed that miR-384 facilitated apoptosis and autophagy of NSCLC cells via repressing the expression of the collagen α-1 (X) chain gene.15 Thus, miR-384 plays a vital role in NSCLC development.
Tripartite motif containing protein 44 (TRIM44) possesses a zinc figure ubiquitin protease domain and exerts function through activating AKT/mTOR pathway.16 Present evidence demonstrated that TRIM44 participated in a variety of human cancers, including cervical cancer,17 glioblastoma,18 ovarian cancer,19 and esophageal cancer.20 In NSCLC, TRIM44 promoted cell growth by regulating NF-κB and was facilitated in NSCLC.21 Therefore, TRIM44 is crucial for NSCLC development. However, the association between the TRIM44 mechanism and NSCLC development is not fully understood.
Here, we detected the expression of circ_0020123, miR-384, and TRIM44 in NSCLC tissues and cells. The effects and underlying mechanism of circ_0020123, miR-384, and TRIM44 on NSCLC cell progression were also assessed. Besides, the role of circ_0020123 in tumor growth was explored in vivo.
Materials and Methods
Tissues Samples Collection and Cell Culture
Thirty-three pairs of NSCLC tissues and corresponding normal tissues were provided by NSCLC patients at Linyi People’s Hospital. The tissue specimens were stored at −80°C for subsequent use. The research was conducted in accordance with the Declaration of Helsinki and was ratified by the Ethics Review Committees of Linyi People’s Hospital. All participants in this research signed informed content.
Two NSCLC cell lines, including adenocarcinoma cell line A549 and large cell lung cancer cell line H1581 and normal cell line (16HBE) were provided by the Cell Bank of the Chinese Academy of Medical Sciences (Beijing, China). All cells were incubated in RPMI-1640/DMEM medium (Gibco, El Paso, TX, USA) with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) at 37°C with 5% CO2.
RT-qPCR Analysis
Total RNA in NSCLC tissues, cells, or tumors was obtained with TRIzol reagent (Invitrogen), followed by DNase treatment to eliminate DNA contamination. The integrity of the RNA in all samples was verified by comparing the ribosomal RNA bands in ethidium bromide-stained gels. RNA sample purity was assessed by using spectrophotometric measurements at 260 and 280 nm. The OD 260/280 of all samples was 1.8–2.2. Then, random primers (Takara, Tokyo, Japan) and PrimeScript™ RT Master Mix Kit (Takara) were applied to reverse transcription circ_0020123, miR-384, and TRIM44. In short, the transcription was carried out in a 10 μL reaction mixture, including polyadenylated RNA (100 ng), 5 × PrimeScript Buffer (2 μL), PrimeScript RT Enzyme Mix I (0.5 μL), RT primer mixture (1 μL), and RNase-free water. Subsequently, 2 μL of cDNA templates and SYBR Green qPCR master mix (Invitrogen) were applied to conduct RT-qPCR. U6 and GAPDH were used as internal loads. The 2−ΔΔCt method was employed to analyze gene expression. The primer sequences of RT-qPCR used in the present study are shown in Table 1.
 |
Table 1 The Primer Sequences of RT-qPCR Used in the Present Study |
RNase R Digestion
For exploring the resistance of circRNA to the RNase R exonuclease digestion. Total RNAs (2 μg) were treated with or without RNase R (Thermo, USA) at 37 °C for 15 min. Following, the expression of circ_0020123 and linear PDZD8 was detected by RT-qPCR assay.
Cell Transfection
Small interfering RNA against circ_0020123 (si-circ_0020123), small hairpin RNA against circ_0020123 (sh-circ_0020123), miR-384 mimic (miR-384), miR-384 inhibitor (anti-miR-384), si-TRIM44, and their negative controls (si-NC, sh-NC, miR-NC, and anti-miR-NC) were obtained from GenePharma (Shanghai, China). For overexpression of circ_0020123, full-length of its sequence was cloned into the pcDNA3.1 vector (GenePharma). Cell transfection was carried out using Lipofectamine 3000 (Invitrogen).
Cell Viability Assay
Briefly, H1581 or A549 cells were transfected and cultured for 0 h, 24 h, 48 h, or 72 h. Then, 20 μL MTT solution (5 mg/mL; Promega, Madison, WI, USA) was added to cell-culture medium for 4 h. Next, the product was dissolved with 200 μL dimethyl sulfoxide (DMSO). Finally, the optical density (OD) of samples was measured using a microplate reader (Bio-Rad, Richmond, CA, USA) at 490 nm.
Cell Apoptosis Assay
Cells were resuspended with binding buffer and stained by Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) (Vazyme Biotech, Nanjing, China). Then, a flow cytometer (ACEA, Agilent, Santa Clara, USA) was used to analyze the apoptotic rate of NSCLC cells.
Cell Migration and Invasion Assay
A transwell chamber (BD Biosciences) was employed to determine the number of migrated and invaded NSCLC cells. The Transwell compartment was coated with Matrigel for invasion assay. 100 μL serum-free medium with the cells and 600 μL corresponding medium with 10% FBS were used for the upper and lower chamber, respectively. After 24 h of incubation, the cells on the upper chamber were discarded using a cotton swab. Cells attached to the lower surface were fixed using methanol for 30 min and stained with 0.1% crystal violet solution. Then, average cell numbers in 10 randomly selected fields were counted by a microscope at×100 magnification.
Western Blot
Total proteins in NSCLC tissues and cells were isolated using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China). Then, the proteins were exposed to 10% SDS-PAGE and then were transferred onto PVDF membrane. Five percent skim milk in phosphate-buffered solution (PBST) was used to block the membrane. Next, the primary antibodies against TRIM44 (1:1000, Abcam, Cambridge, MA, USA), E-cadherin (1:1000, Abcam), N-cadherin (1:1000, Abcam), Vimentin (1:1000, Abcam), or GAPDH (1:1000, Abcam) and corresponding secondary antibody Goat Anti-Rabbit IgG H&L (HRP) (1:20,000; Abcam) were used to incubate the membranes. Finally, the protein bands were imaged by a BeyoECL Plus Kit (Beyotime Biotechnology).
Dual-Luciferase Reporter Assay
Firstly, the sequences contain wild (WT) or mutant (MUT) type binding sites of circ_0020123 or TRIM44 3ʹuntranslated region (UTR) in miR-384 was cloned into the pGL3 vector (Promega). Then, H1581 and A549 cells were co-transfected with the reporter gene plasmids and miR-484 or miR-NC. After incubation for 24 hours, a Dual Luciferase Reporter Assay System (Promega) was employed to determine the luciferase intensity based on the manufacturer’s instruction.
RNA Pull-Down Assay
H1581 and A549 cells were transfected with biotinylated (Bio)-TRIM44-WT, Bio-TRIM44-MUT, or Bio-NC (a negative control) (Genescript, Nanjing, China) at a final concentration of 100 nM. At 48 h after transfection, cells were harvested and sonicated. Remaining cell lysates were incubated with streptavidin-coated magnetic beads (Invitrogen) at 4 °C for 2 h and then purified using RNeasy Mini Kit (QIAGEN, Duesseldorf, Germany) for analysis. Finally, the RT-qPCR assay was applied to detect RNA enrichment. Ranilla luciferase was used as an internal control.
RNA Immunoprecipitation
RNA immunoprecipitation (RIP) assays were employed with EZ-Magna Nuclear RIP (Cross-Linked) RNA-Binding Protein Immunoprecipitation Kit (Sigma, Merck KGaA, Darmstadt, Germany) following the manufacturer’s protocol. Briefly, NSCLC cells were lyzed in RIP lysis buffer. Then 100 μL of whole cell lysate was co-incubated with RIP buffer including magnetic beads conjugated with human antibody against Argonaute2 (Ago2) (1:50, Abcam) or normal mouse IgG (1:25, Abcam) for 6 h at 4°C. Then the beads were treated with Proteinase K buffer and the target RNA was extracted. Co-precipitated RNAs were finally subjected to RT-qPCR analysis.
Mouse Xenografts
This experiment was carried out in accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of china) and was authorized by the Animal Research Committee of Linyi People’s Hospital. Briefly, 5-week-old BALB/c nude mice were subcutaneously injected with A549 cells stably transfected with sh-NC or sh-circ_0020123, each group consisted of 7 mice. After injection for 7 days, the tumor volume (length × width2/2) was measure every 7 days. After injection for 4 weeks, the tumors were collected for the analysis of weight and further experiment.
Statistical Analysis
All data were shown as the mean ± standard deviation (SD). Student’s t-test was utilized to analyze the data between two groups. Spearman correlation coefficient was employed to investigate the association between levels of circ_0020123 and TRIM44, circ_0020123 and miR-384, or TRIM44 and miR-384. A Kaplan-Meier curve and the Log rank test were employed for survival analysis. P < 0.05 was considered as statistical significance.
Results
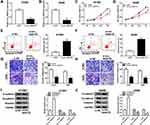
circ_0020123 Expression Was Elevated, and miR-384 Expression Was Repressed in NSCLC Tissues and Cells
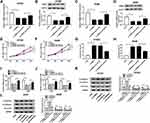
The expression of circ_0020123 and miR-384 in NSCLC tissues and cells was first assessed by RT-qPCR. Our result showed that circ_0020123 expression was obviously increased in NSCLC tissues and cells in contrast with the normal tissues and cells (Figure 1A and B). However, miR-384 expression level in NSCLC tissues and cells was remarkably decreased in NSCLC cells (Figure 1C and D). We further disclosed the relationship between circ_0020123 and miR-384 expressions in NSCLC tissues. As displayed in Figure 1E, the miR-384 expression was negatively correlated with circ_0020123 level. The high expressed circ_0020123 was also negatively correlated with the overall survival rate of patients (Figure 1F). Besides, circRNA exhibits natural resistance to exonucleases due to its high stability. Thus, we detected the resistance of circ_0020123 to RNase R. As displayed in Figure 1G, RNase R treatment significantly reduced the expression of linear PDZD8 while had little effect on circ_0020123 expression. Therefore, circ_0020123 was a circular RNA and its expression negatively relevant to miR-384 expression.
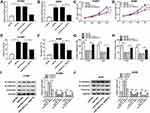
circ_0020123 Knockdown Inhibited the Growth of NSCLC Cells
We further confirmed the effect of circ_0020123 on NSCLC development. As displayed in Figure 2A and B, the transfection with si-circ_0020123 strongly downregulated the expression of circ_0020123. The MTT results suggested that cell viability was repressed by circ_0020123 knockdown in H1581 and A549 cells (Figure 2C and D). In addition, circ_0020123 depletion promoted the apoptosis of H1581 and A549 cells (Figure 2E and F). Besides, the cell migration and invasion abilities were remarkably inhibited in cells transfected with si-circ_0020123 compared with the cells with si-NC transfection (Figure 2G and H). And it was further confirmed by the elevated protein level of epithelial-mesenchymal transitions (EMT) marker E-cadherin, and the decreased protein levels of N-cadherin and Vimentin in H1581 and A549 cells (Figure 2I and J). These data showed that circ_0020123 knockdown suppressed NSCLC progression.
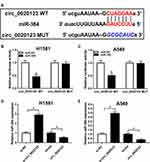
miR-384 Was a Target Gene of circ_0020123
Ample research has indicated that circRNAs acted as a competing endogenous RNAs for miRNAs. Therefore, we were curious that if circ_0020123 contains the binding sites to miR-384. Bioinformatics analysis tool starbase showed that circ_0020123 contained the binding sites of miR-384 (Figure 3A). Moreover, this correlation was further confirmed by dual-luciferase reporter assay. Transfection of miR-384 significantly reduced the luciferase activity of circ_0020123 WT, while it has little change in circ_0020123 MUT group (Figure 3B and C). Besides, si-circ_0020123 and circ_0020123 overexpression plasmid was employed and RT-qPCR assay was conducted to examine the changes on miR-384 expression. As indicated in Figure 3D and E, the level of miR-384 was dramatically upregulated by circ_0020123 knockdown and downregulated by circ_0020123 overexpression. Thus, circ_0020123 targeted to miR-384 and negatively modulated its expression.
circ_0020123 Regulated NSCLC Cell Progression by miR-384
As miR-384 was a target of circ_0020123, we subsequently detected the effect of miR-384 on circ_0020123-mediated NSCLC progression. H1581 and A549 cells were transfected with miR-NC, miR-384, miR-384 + vector, or miR-384 + circ_0020123, respectively. QRT-PCR assay suggested that miR-384 level was increased by miR-384 overexpression, but it was reversed by circ_0020123 overexpression (Figure 4A and B). As shown in Figure 4C and D, cell viability was suppressed by miR-384 upregulation, and then rescued by circ_0020123 overexpression. Moreover, flow cytometry analysis showed that increased circ_0020123 expression inhibited cell apoptosis promoted by a miR-384 upregulation in H1581 and A549 cells (Figure 4E and F). Next, we found that miR-384 overexpression significantly repressed cell migration and invasion abilities, whereas this effect was alleviated by the upregulation of circ_0020123 (Figure 4G and H). Besides, we also found that the upregulation of circ_0020123 reversed the effect of miR-384 overexpression on the levels of EMT-related proteins (Figure 4I and J). These data demonstrated that circ_0020123 regulated the growth of NSCLC cells by downregulating miR-384.
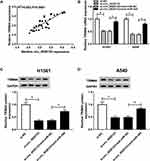
miR-384 Targeted TRIM44 and Downregulated TRIM44 Expression
Based on the above results, we next investigated the downstream genes of miR-384 using the bioinformatics tool targetscan, and found that TRIM44 possessed a motif with complementary sequence to miR-384 (Figure 5A). The luciferase activity of TRIM44 3ʹUTR WT, but not TRIM44 3ʹUTR MUT, was attenuated by miR-384, indicating interaction between the miR-384 and TRIM44 (Figure 5B and C). To further verify the direct correlation between miR-384 and TRIM44, RNA pull-down assay and RIP assay were carried out in NSCLC cells. The result suggested that the enrichment of miR-384 in the Bio-TRIM44-WT group was apparently increased in contrast with the Bio-TRIM44-MUT group and Bio-NC group in H1581 and A549 cells (Figure 5D and E). As displayed in Supplementary Figure 1A and B, both miR-384 and TRIM44 were significantly enriched in the Ago2 group in contrast with the IgG immunoprecipitates. Subsequently, we analyzed the mRNA and protein levels of TRIM44 in NSCLC tissues and cells. The mRNA and protein levels of TRIM44 was significantly increased in NSCLC tissues and cells (Figure 5F–I). Furthermore, we investigated whether miR-384 regulating TRIM44 expression. As displayed in Figure 5J–L, the mRNA level and protein level of TRIM44 was remarkably reduced by miR-384 upregulation and dramatically increased by miR-384 depletion. In addition, TRIM44 expression was inversely related to miR-384 expression in NSCLC tissues (Figure 5M). Taken together, miR-384 downregulated TRIM44 expression via targeting TRIM44.
miR-384 Mediated TRIM44 Expression to Regulate NSCLC Progression
For exploration of the effect of TRIM44 on NSCLC progression, H1581 and A549 cells were transfected with si-NC, si-TRIM44, si-TRIM44 + anti-miR-NC, or si-TRIM44 + anti-miR-384. TRIM44 mRNA and protein levels were downregulated by si-TRIM44, and partly restored by miR-384 depletion (Figure 6A–D). Besides, we found that TRIM44 downregulation repressed cell viability, whereas this effect was reversed due to the depletion of miR-384 (Figure 6E and F). Moreover, flow cytometry analysis suggested that miR-384 knockdown inhibited cell apoptosis induced by TRIM44 depletion in H1581 and A549 cells (Figure 6G and H). Furthermore, cell migration and invasion abilities suppressed by TRIM44 depletion were induced by miR-384 knockdown (Figure 6I and J, Supplementary Figure 2A and B). Besides, the reversion effect of miR-384 depletion on the progression by which TRIM44 knockdown regulating EMT-related protein level was discovered in H1581 and A549 cells (Figure 6K and L). Taken together, miR-384 regulated NSCLC cell progression by suppressing TRIM44 expression.
circ_0020123 Suppressed miR-384 Expression to Upregulate TRIM44 Level
Based on the above findings, we hypothesized that circ_0020123 targets to miR-384 to regulate TRIM44 expression. As expected, there was a positive correlation between circ_0020123 and TRIM44 expressions in NSCLC tissues (Figure 7A). Besides, H1581 and A549 cells were transfected with si-circ_0020123 + anti-miR-384, si-circ_0020123 + anti-miR-NC, si-circ_0020123 or si-NC, respectively. The data demonstrated that TRIM44 expression was downregulated by circ_0020123 knockdown, and then upregulated by miR-384 depletion (Figure 7B–D). Therefore, circ_0020123 regulates the TRIM44 expression by targeting miR-384.
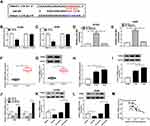
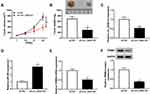
circ_0020123 Knockdown Repressed Tumor Growth in vivo
To investigate whether circ_0020123 affecting tumor growth in vivo, A549 cells with stable circ_0020123 knockdown were established. Then, BALB/c nude mice were subcutaneously injected with A549/sh-NC or A549/sh-circ_0020123. As displayed in Figure 8A and B, circ_0020123 depletion significantly inhibited the tumor volume and tumor weight. Besides, the expressions of circ_0020123, miR-384, and TRIM44 were analyzed in transplanted tumors. As expected, circ_0020123 (Figure 8C) and TRIM44 (Figure 8E and F) were downregulated, while miR-384 (Figure 8D) was upregulated in tumors in sh-circ_0020123 group. These data confirmed that circ_0020123 depletion suppressed tumor growth in vivo.
Discussion
CircRNAs have been reported to be related to NSCLC development.22,23 For example, Cui et al confirmed that circ_0043278 was considered a biomarker and induced cell growth and mobility via modulating miR-520f in NSCLC cells.24 Wang et al revealed that circ_0008305 repressed TGF-β-induced EMT by regulating miR-429/miR-200b-3p expression, and circ_0008305 level was dramatically decreased in TGF-β-induced NSCLC cells.25 Zhou et al demonstrated that circ_0004015 mediated NSCLC cell growth and chemo-sensitivity via regulating the miR-1183/PDPK1axis.26 Therefore, circRNAs play pivotal roles in NSCLC cell progression.
In this paper, the elevated circ_0020123 expression was found in NSCLC tissues and cells. Furthermore, circ_0020123 knockdown significantly suppressed the growth of NSCLC cells. In addition, the xenograft model indicated that circ_0020123 depletion could strongly repress tumor growth in vivo. These data were consistent with the previous data suggested that circ_0020123 was increased and acted as a carcinogenic factor in NSCLC cells.10,27 These results indicating that circ_0020123 could serve as a novel biomarker for NSCLC. However, the underlying mechanism of circ_0020123 in human cancers was poorly understood. Thus, more experiments are needed to confirm its function in NSCLC.
Increasing studies have suggested that circRNAs could target to miRNAs and modulate miRNAs expression.28 MiR-384 was identified as a suppressive gene in types of human cancers, such as osteosarcoma,29 prostate cancer,30 gastric cancer,31 and NSCLC.12 Moreover, miR-384, targeted by circPAPPA, affected the development of trophoblast cells.32 Besides, many long non-coding RNAs (lncRNAs), including SNHG3,33 TUG1,34 and NEAT1,35 were revealed to target miR-384 in cancer development. However, the relationship between circ_0020123 and miR-384 in NSCLC remains inconclusive. In this study, we confirmed that miR-384 was a direct target of circ_0020123, and circ_0020123 negatively regulated the miR-384 level in NSCLC cells. Furthermore, decreased miR-384 level was observed in NSCLC tissues and cells. Thus, we hypothesized that circ_0020123 repressed miR-384 expression to regulate the growth of NSCLC cells. Then, we designed the experiments to verify this hypothesis.
The majority of miRNAs are identified as sponges for mRNAs to regulate gene expression.36 TRIM44 was a target of miR-384 and was inhibited by miR-384. Spearman correlation analysis also confirmed that the level of TRIM44 was negatively related to miR-384 expression in NSCLC tissues. Besides, elevated TRIM44 was discovered in NSCLC tissues, which was in line with the previous findings.21 Although present evidence suggested that TRIM44 promoted the growth of NSCLC cells, the functional studies of TRIM44 in NSCLC were poor. In our study, TRIM44 knockdown inhibited cell growth, mobility, EMT, and elevated apoptosis in NSCLC cells. Also, we confirmed that miR-384 affected NSCLC cell progression via repressing TRIM44 expression. These data indicated that TRIM44 played a crucial role in NSCLC development.
Finally, the association between circ_0020123, miR-384, and TRIM44 was explored. As expected, circ_0020123 impeded miR-384 expression to promote TRIM44 expression in NSCLC cells. Previous literature demonstrated that circ_0020123 knockdown inhibited tumor growth of NSCLC in vivo.29 Consistently our results revealed that circ_0020123 depletion repressed cell growth in vivo, including tumor volume and weight.
In summary, our data demonstrated that circ_0020123 knockdown repressed the viability, mobility, and EMT, and induced apoptosis of NSCLC cells, revealing a new mechanism by which circ_0020123 was regulating NSCLC cell progression. Furthermore, our findings provided a potential novel target for the NSCLC therapy.
Highlights
- The interaction between miR-384 and circ_0020123 or TRIM44 was confirmed for the first time
- Circ_0020123 knockdown represses NSCLC cell progression
- Circ_0020123 modulates the progression of NSCLC through regulating miR-384/TRIM44 axis
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi:10.3322/caac.20138
2. DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi:10.3322/caac.21235
3. Hsieh JJ, Hou MM, Chang JW, Shen YC, Cheng HY, Hsu T. RAB38 is a potential prognostic factor for tumor recurrence in non-small cell lung cancer. Oncol Lett. 2019;18:2598–2604.
4. Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi:10.1016/j.molcel.2015.03.027
5. Bolha L, Ravnik-Glavac M. Circular RNAs: biogenesis, function, and a role as possible cancer. Biomarkers. 2017;2017:6218353.
6. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi:10.1016/j.cell.2015.02.014
7. Hao S, Lv J, Yang Q, et al. Identification of key genes and circular RNAs in human gastric cancer. Med Sci Monit. 2019;25:2488–2504. doi:10.12659/MSM.915382
8. Zhang S-J, Chen X, Li C-P, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci. 2017;58(14):6500–6509. doi:10.1167/iovs.17-22698
9. Wang W, Wang Y, Piao H, et al. Circular RNAs as potential biomarkers and therapeutics for cardiovascular disease. PeerJ. 2019;7:e6831.
10. Wan J, Hao L, Zheng X, Li Z. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488–3p to regulate ADAM9 expression. Biochem Biophys Res Commun. 2019;515(2):303–309. doi:10.1016/j.bbrc.2019.05.158
11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
12. Fan N, Zhang J, Cheng C, Zhang X, Feng J, Kong R. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling. Biomed Pharmacother. 2017;95:1331–1337. doi:10.1016/j.biopha.2017.08.143
13. Li J, Chen M, Yu B. miR-433 suppresses tumor progression via Smad2 in non-small cell lung cancer. Pathol Res Pract. 2019;215(10):152591. doi:10.1016/j.prp.2019.152591
14. Iacona JR, Monteleone NJ. Transcriptomic studies provide insights into the tumor suppressive role of miR-146a-5p in non-small cell lung cancer (NSCLC). cells. 2019;1–12.
15. Guo Q, Zheng M. MiR-384 induces apoptosis and autophagy of non-small cell lung cancer cells through the negative regulation of Collagen alpha-1(X) chain gene. 2019;39.
16. Wei CY, Wang L, Zhu MX, et al. TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J Exp Clin Cancer Res. 2019;38:137. doi:10.1186/s13046-019-1138-7
17. Liu S, Meng F, Ding J, et al. High TRIM44 expression as a valuable biomarker for diagnosis and prognosis in cervical cancer. Biosci Rep. 2019;39.
18. Li L, Shao MY, Zou SC, Xiao ZF, Chen ZC. MiR-101-3p inhibits EMT to attenuate proliferation and metastasis in glioblastoma by targeting TRIM44. J Neuro-Oncol. 2019;141:19–30.
19. Liu S, Yin H, Ji H, Zhu J, Ma R. Overexpression of TRIM44 is an independent marker for predicting poor prognosis in epithelial ovarian cancer. Exp Ther Med. 2018;16:3034–3040.
20. Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018;109:3080–3092.
21. Luo Q, Lin H, Ye X, Huang J, Lu S, Xu L. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF-kappaB signaling pathway. Int J Clin Oncol. 2015;20:508–517. doi:10.1007/s10147-014-0752-9
22. Liu YT, Han XH, Xing PY, et al. Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J Thorac Dis. 2019;11:1779–1787. doi:10.21037/jtd.2019.05.22
23. Chen T, Luo J. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer. 2019;10:930–941.
24. Cui J, Li W, Liu G, et al. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47:810–821. doi:10.1080/21691401.2019.1575847
25. Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17:140.
26. Zhou Y, Zheng X, Xu B, et al. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys Res Commun. 2019;508:527–535. doi:10.1016/j.bbrc.2018.11.157
27. Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8:1387–1402.
28. Piwecka M, Glazar P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357.
29. Wang Y, Huang H, Li Y. Knocking down miR-384 promotes growth and metastasis of osteosarcoma MG63 cells by targeting SLBP. Artif Cells Nanomed Biotechnol. 2019;47(1):1458–1465. doi:10.1080/21691401.2019.1601099
30. Hong Z, Fu W, Wang Q, Zeng Y, Qi L. MicroRNA-384 is lowly expressed in human prostate cancer cells and has anti-tumor functions by acting on HOXB7. Biomed Pharmacother. 2019;114:108822. doi:10.1016/j.biopha.2019.108822
31. Wang F. miR-384 targets metadherin gene to suppress growth, migration, and invasion of gastric cancer cells. J Int Med Res. 2019;47(2):926–935. doi:10.1177/0300060518817171
32. Zhou W, Wang H, Yang J, et al. Down-regulated circPAPPA suppresses the proliferation and invasion of trophoblast cells via the miR-384/STAT3 pathway. Biosci Rep. 2019;39(9). doi:10.1042/BSR20191965
33. Wang L, Su K, Wu H, Li J, Song D. LncRNA SNHG3 regulates laryngeal carcinoma proliferation and migration by modulating the miR-384/WEE1 axis. Life Sci. 2019;232:116597. doi:10.1016/j.lfs.2019.116597
34. Qian W, Ren Z, Lu X. Knockdown of long non-coding RNA TUG1 suppresses nasopharyngeal carcinoma progression by inhibiting epithelial-mesenchymal transition (EMT) via the promotion of miR-384. Biochem Biophys Res Commun. 2019;509(1):56–63. doi:10.1016/j.bbrc.2018.12.011
35. Zhu L, Yang N, Li C, Liu G, Pan W, Li X. Long noncoding RNA NEAT1 promotes cell proliferation, migration, and invasion in hepatocellular carcinoma through interacting with miR-384. J Cell Biochem. 2018.
36. Doench JG. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–511. doi:10.1101/gad.1184404
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.