Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA circ_0001105 Inhibits Progression and Metastasis of Osteosarcoma by Sponging miR-766 and Activating YTHDF2 Expression
Authors Yang J, Han Q, Li C, Yang H, Chen X, Wang X
Received 15 October 2019
Accepted for publication 10 January 2020
Published 26 February 2020 Volume 2020:13 Pages 1723—1736
DOI https://doi.org/10.2147/OTT.S234668
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Jie Yang, 1,* Qicai Han, 2,* Chao Li, 3 Hao Yang, 2 Xiaolong Chen, 2 Xiaohu Wang 1
1Department of Orthopedics, Zhengzhou Central Affiliated Hospital to Zhengzhou University, Zhengzhou 450052, People’s Republic of China; 2Department of Orthopedics, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China; 3Department of Bone and Soft Tissue, The Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaohu Wang
Department of Orthopedics, Zhengzhou Central Affiliated Hospital to Zhengzhou University, Zhengzhou 450052, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) play vital roles in the modulation of tumor progression. This study explored the biological functions of circ_0001105 in the progression of osteosarcoma (OS).
Methods: qRT-PCR and in situ hybridization (ISH) were performed to detect the expression status of circ_0001105 in cells and tissues. Bioinformatics analysis, dual-luciferase reporter gene assay, Western blot and qRT-PCR were performed to determine the relationships among RNAs. The CCK-8, colony formation, EdU, transwell and wound healing assays were conducted to evaluate the cell growth, invasion and migration of OS cells. Tumor xenografts were established to investigate the effects of circ_0001105 on tumor growth in vivo. Lastly, the protein expression of YTHDF2 in OS tissues was measured using immunohistochemical staining.
Results: Data showed that circ_0001105 and YTHDF2 were significantly lower, while miR-766 was higher in OS tissues compared to adjacent tissues. Low expression of circ_0001105 or YTHDF2 was associated with poor survival of OS patients as demonstrated by the Kaplan-Meier analysis. In addition, miR-766 was identified as a direct binding target of circ_0001105 and YTHDF2. Ectopic overexpression of circ_0001105 or YTHDF2 significantly suppressed OS cell viability and invasion through regulating miR-766. Last, overexpression of circ_0001105 significantly attenuated in vivo tumor growth.
Conclusion: Our findings suggest that circ_0001105 inhibits OS progression, at least partially, by regulating miR-766/YTHDF2 signaling pathway.
Keywords: circ_0001105, OS, progression, miR-766, YTHDF2
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor posing the biggest health threats to children and young adolescents.1,2 More recently, the emergence of neoadjuvant combined with cooperative treatment has dramatically improved the prognosis of localized OS. However, the prognosis of patients with advanced OS metastatic or recurrent status remains poor.3 In light of this, to improve therapeutic treatment and prognostic outcome(s), a comprehensive understanding of the genetic changes associated with OS progression is vital.
Non-coding RNAs are a highly heterogenous group of protein-coding RNAs consisting of: (i) microRNAs (miRNAs), less than 200 nt in length; (ii) long non-coding RNAs (lncRNAs), longer than 200 nt and more recently; (iii) circular RNAs (circRNAs).4–6 A growing body of knowledge indicates that circRNAs are differentially expressed in a variety of human cancers and play important roles in the regulation of gene expression by sponging miRNAs.7 Unlike lncRNAs and miRNAs, circRNAs are evolutionally conserved, stable and enormously abundant in cells.5 These features make circRNAs potential biomarkers or therapeutic targets for various human diseases, especially cancers. In addition, circRNAs have been found to be involved in the tumorigenesis and the development of neoplasms, such as esophageal squamous cell carcinoma (circRNA_100876 and circ-DLG1),8,9 pancreatic carcinoma (circRNA_100782),10 breast cancer (circ_0008945),11 lung cancer (circ_0023404, circ-SMARCA5),12,13 hepatocellular carcinoma (circ_0003998),14 and OS (circ_0001721, circ-LRP6).15,16 However, the potential roles of circRNAs in OS are yet to be fully documented yet.
This study identified a novel dysregulated expression circRNA, circ_0001105, in osteosarcoma. So far, the expression status of circ_0001105 in OS has not been explored. Hence, we analyzed the expression of circ_0001105 and characterized its correlation with the prognosis of OS patients. Likewise, the effect of circ_0001105 on the malignant biological behavior of osteosarcoma was investigated via circ_0001105/miR-766/YTHDF2 signaling pathway (Supplementary Figure 1).
Materials and Methods
Human Specimens
This study was approved and supervised by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, Zhengzhou Central Affiliated Hospital to Zhengzhou University, and The Affiliated Cancer Hospital of Zhengzhou University. In total, 120 tissue specimens and 65 surrounding non-tumorous tissues were collected from OS patients at the above-mentioned hospitals. All the patients received primary surgical treatment and preoperative/postoperative neoadjuvant therapy. Written informed consent was obtained from all patients for research purposes and ethical approval for the study was obtained from the Ethics Committee of Zhengzhou University (protocol number: OSRE-2011-082). Clinical information and follow-up records were obtained through medical records and mobile telephones. Good or poor chemotherapy response was distinguished based on a 90% necrosis cutoff. Detailed clinicopathological parameters of the 120 patients are summarized in Table 1 and Supplementary Table 1.
 |
Table 1 Correlation of Clinico-Pathological Features with circRNA-0001105 Expression in Osteosarcoma Cohort |
Cell Culture
MG63, U2OS, and 143b were purchased from the American Type Culture Collection (Rockville, USA) while HOBC and HFOB were purchased from Shanghai Institute for Biological Science (Shanghai, China). Cell lines were routinely grown in suitable media as recommended and maintained at 37°C in an incubation cabinet containing 5% CO2.
Cell Transfection and Plasmid Constructs
The miR-766 mimics, anti-miR-766 or miR-NC, shRNAs targeting circ_0001105 were synthesized by Hanbio (Shanghai, China) and transfected into MG63 and U2OS cells using the LipoFiter (Shanghai, China) according to the manufacturer’s instructions. RNAi sequences used in the study are all listed in Supplementary Table 2 and 3. The YTHDF2 human sequence was obtained from reverse transcription (RT) of total RNA.
Immunohistochemistry (IHC) and in situ Hybridization (ISH) Staining
Stained paraffin-embedded immunohistochemical tissue sections were processed as previously described.17 Primary antibodies used in the present study are listed as follows: YTHDF2 (24744-1-AP, Proteintech, Wuhan, China), Ki-67 (9449, Cell Signaling Technology, MA, USA). Three areas were chosen randomly from each section for measurement. We employed in situ hybridization to investigate the expression and intracellular location of circ_0001105 in OS tissues as previously described.18,19
Western Blotting
Western blotting was processed as previously described.20 Primary antibodies used in the present study are listed as follows: YTHDF2 (24744-1-AP, Proteintech), GAPDH (9449, Cell Signaling Technology).
CCK-8, Colony Formation, Cell Invasion and Migration Assay
The CCK-8, colony formation, cell invasion and migration assay were performed according to the previously described method.17
Dual-Luciferase Reporter Gene Assay
Dual-luciferase reporter gene assay was processed as previously described.20 Luciferase activities were measured with a dual-luciferase reporter assay system (Promega, WI, USA).
In vivo Experiments
Animal experiments were conducted as described previously.21 To establish the nude mice models of OS, we injected U2OS cells with control shRNA (NC) or shRNA targeting circ_0001105 (sh-circ_0001105) into the flank of the nude mice. Tumor volume, tumor weight and luciferase signal were measured. The expression of Ki-67 was measured with IHC assay. This animal experiment was approved by the Animal Ethics Committee of the First Affiliated Hospital of Zhengzhou University. In addition, Guide for the Care and Use of Laboratory Animals (8th edition) was strictly followed in the present study.
Statistical Analysis
All data are expressed as mean ± SEM. The significant difference was determined by two-tailed Student’s t test, ANOVA test, Chi-square test, Pearson correlation and Kaplan-Meier analysis as appropriate. P<0.05 was considered statistically significant.
Results
circ_0001105 Is Correlated with Tumor Metastasis and Survival of OS Patients
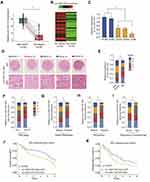
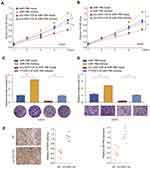
The relative expression level of circ_0001105 was measured in 30 patients with OS and surrounding non-tumorous tissues using qRT-PCR. Notably, circ_0001105 was remarkedly upregulated in OS tissue (Figure 1A and B). Meanwhile, circ_0001105 level was also higher in OS cell lines than that seen in normal human osteoblast cell lines (Figure 1C). Subsequently, we determined circ_0001105 expression in OS TMA using the ISH assay, and the results revealed the elevated circ_0001105 expression level in OS (Figure 1D and E). In addition, circ_0001105 was observably highly expressed in advanced clinical TNM stage OS tumors (Figure 1F). Additionally, circ_0001105 expression was higher in metastatic tumors (Figure 1G), recurrent tumors (Figure 1H) and was associated with poor response to chemotherapy (Figure 1I) OS tissues. To further understand the significance of circ_0001105 in OS, we determined the potential associations between circ_0001105 expression and the patients’ clinicopathological features in cohort of OS TMA cohort containing 120 OS tissues. The 120 OS patients were classified into two groups based on the circ_0001105 ISH staining intensive: (i) high-circ_0001105 group (n=57), and (ii) low-circ_0001105 group (n=63). Kaplan-Meier analysis showed that a lower circ_0001105 expression in the tumor tends to confer a significantly poor prognosis (Figure 1J and K). Moreover, univariate and multivariate Cox proportional hazards analyses confirmed that circ_0001105 level was an independent prognostic factor in patients with OS (Table 2). These findings indicated that low expression of circ_0001105 predicted favorable prognosis of patients with OS.
 |
Table 2 Correlation of Clinic-Pathological Features with circRNA-0001105 Expression in OS Cohort |
The Effect of circ_0001105 on the Regulation of OS Cell Proliferation and Invasion
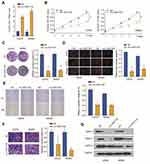
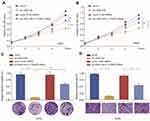
To investigate the function of circ_0001105 in OS cells, we, respectively, carried out gain-function experiments by overexpression circ_0001105 in OS cells (Figure 2A). The ectopic overexpression of circ_0001105 significantly reduced cell proliferation in U2OS and MG63 cell lines as revealed by the CCK-8 assays and colony formation assay (Figure 2B and C). Overexpression of circ_0001105 suppressed the DNA synthesis rate when compared with the negative control (Figure 2D). Furthermore, cell migration and transwell assays showed that overexpression of circ_0001105 significantly decreased the migration and invasion of U2OS and MG63 cells compared with the NC groups, respectively (Figure 2E and F). Overexpression of circ_0001105 also decreased the levels of the metastasis-associated protein MMP-2, MMP-7 and MMP-9 (Figure 2G). These results indicate that circ_0001105 regulates the proliferation and invasion of OS cells.
circ_0001105 Knockdown Suppresses Tumorigenesis in a Xenograft Model
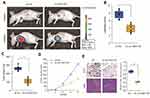
Based on the in vitro results, we postulated that circ_0001105 may suppress tumor growth in animals. To test this hypothesis, U2OS cells stably overexpressing circ_0001105 or negative control cells were subcutaneously injected into the flanks of nude mice. As predicted, we observed that the mice in circ_0001105 overexpression had decreased luciferase photon flux than the negative control group (Figure 3A and B). Moreover, tumors extracted from mice injected with cells overexpressing circ_0001105 were lighter than mice in the control group (Figure 3C). Meanwhile, the rate of tumor growth was slower in the circ_0001105 overexpression group than in the control group (Figure 3D). Furthermore, overexpression of circ_0001105 showed lower Ki-67 IHC staining intensity in the xenografted tissues relative to the control group (Figure 3E). The above results in vitro and in vivo results further illustrate the crucial role of circ_0001105 in OS cell growth and invasion. However, the exact underlying mechanisms remain unclear.
circ_0001105 Is as a miRNA Sponge for the miR-766
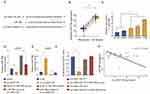
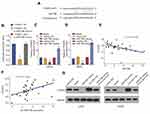
Given that circRNA acts as a miRNA sponge, we explored whether circ_0001105 could bind to miRNAs. The binding sequences between circ_0001105 and miR-766 were predicted using online tools (https://circinteractome.nia.nih.gov/) (Figure 4A). We also found that miR-766 was evidently up-regulated in OS tissues and cell lines (Figure 4B and C). Additional, as shown in Figure 4D and E, overexpression of circ_0001105 significantly inhibited miR-766 expression, while circ_0001105-depletion dramatically increased miR-766 expression level. Moreover, the direct interaction between miR-766 and circ_0001105 was further corroborated through the luciferase reporter assay (Figure 4F). Pearson correlation analysis suggested that circ_0001105 was negatively correlated with miR-766 expression in OS tissues (Figure 4G).
To further clarify whether miR-766 was directly involved in the circ_0001105-mediated anti-proliferation effect of OS, U2OS stably overexpressing circ_0001105 were transiently transfected with miR-766 mimics. The cell viability and colony formation ability of U2OS in the miR-766 mimics group were significantly increased, while induction of circ_0001105 could abolish the role of miR-766 in promoting cell growth (Figure 5A–C). Consistently, the enhanced cell invasion following overexpression of miR-766 was reversed by co-expression of circ_0001105 (Figure 5D–E). Together, these data indicated that circ_0001105 suppresses OS progression by targeting miR-766.
YTHDF2 as the Downstream Functional Target Gene of circ_0001105/miR-766 Axis
Online tools (http://starbase.sysu.edu.cn) were utilized to identify the targets of miR-766, and we placed emphasis on the YTHDF2 (Figure 6A). First, dual-luciferase reporter assay was conducted to confirm the target relationship between YTHDF2 and (Figure 6B). Moreover, YTHDF2 expressions were detected using qRT-PCR after altering miR-766 expression in U2OS and SAOS cells. Interestingly, the mRNA levels of YTHDF2 were significantly decreased in miR-766 overexpression group but were downregulated in the miR-766 knockdown group (Figure 6C and D). Additionally, Pearson analysis showed a negative correlation between YTHDF2 and miR-766, but a positive correlation between YTHDF2 and circ_0001105 in OS samples (Figure 6E and F). Furthermore, Western blot assay showed that protein levels of YTHDF2 were significantly increased in the circ_0001105 overexpression group but were downregulated when transfected with miR-766 mimic. The. co-transfected with circ_0001105 and miR-766 mimic reversed the inhibitory effects of miR-766 on YTHDF2 expression (Figure 6E and F).
To further figure out the role of circ_0001105/miR-766/YTHDF2 axis in tumor progress, we knockdown YTHDF2 expression in circ_0001105-overexpressed U2OS and SAOS cells. Functional experiments revealed that YTHDF2 silencing significantly rescued OS cell proliferation invasion impaired by circ_0001105 overexpression (Figure 7A and B). Additionally, we checked the expression levels of YTHDF2 and miR-766 in tumor tissues from nude mice injected with cells overexpressing circ_0001105. We found that the levels of YTHDF2 were higher in tumor tissues of the circ_0001105 overexpression group than that in the control group, while the miR-766 level was lower (Figure 7C). Moreover, IHC analysis also indicated that there were higher protein levels of YTHDF2 in tumor tissues of the circ_0001105 overexpression group (Figure 7D). Taken together, our results demonstrated that circ_0001105 inhibitor OS cell proliferation and invasion in a miR-766/YTHDF2 axis-dependent manner.
Correlation Between YTHDF2 Expression and Clinical Characteristics of OS
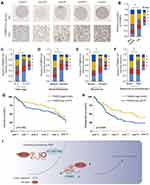
Pan-cancer analysis based on the TCGA database showed that YTHDF2 expression is frequently dysregulated in various cancers (Supplementary Figure 2). To further determine the expression status and prognostic value of YTHDF2 in OS, the relationship between YTHDF2 expression level and the clinicopathological characteristics was analyzed using a TMA cohort. The results showed that YTHDF2 was significantly downregulated in OS tissues (Figure 8A and B). In addition, low expression of YTHDF2 was significantly correlated with clinically aggressive tumor phenotypes, such as advanced TNM stage (Figure 8C), high rate of distance metastasis and recurrence (Figure 8D and E), and poor response to chemotherapy (Figure 8F). Finally, Kaplan-Meier analysis indicated that low expression of YTHDF2 was associated with the worse prognosis of OS patients (Figure 8G and H). In summary, these findings provide evidence that YTHDF2 is involved in the tumorigenesis and progression of OS.
Discussion
In the current study, we show that circ_0001105 expression level is significantly decreased in OS tissue and cell lines in comparison with normal tissues. In addition, our data reveal that decreased circ_0001105 expression is closely associated with aggressive malignant phenotypes of OS including larger tumor size, advanced TNM stage and worse prognosis. Moreover, downregulated-circ_0001105 served as an independent prognostic factor for OS patients. These findings demonstrate that the circ_0001105 might act as a tumor suppressor and contribute to the progression of OS. Increasing evidence had revealed that circRNAs could act as oncogenes or tumor suppressors in diverse cancers. For example, circ_009755 might act as tumor suppressor in the pathogenesis of oral squamous cell carcinoma.22 Circ_0006168 and circRNA_100876 might act as oncogene in esophageal squamous cell carcinoma.9,23 Studies have also shown that circ_0001721, circRNA LRP6, circTADA2A, circ_HIPK3, circRNA_100876 and circTADA2A are all aberrantly in osteosarcoma and contribute to cell proliferation, metastasis and poor prognosis, suggesting their oncogene roles in OS.15,16,24–26
Given that circ_0001105 is downregulated in OS tissues, we tested the role of circ_0001105 in OS cells via gain-of-function in vitro and in vivo experiments. As we expected, cell proliferation and invasion potentials were strikingly impeded following circ_0001105 overexpression, indicating that circ_0001105 was a tumor suppressor in OS cells. Similarly, up-regulation of circ_0001105 significantly suppressed tumor growth in OS xenograft model. Overall, circ_0001105 might act as a tumor suppressor and contribute to the tumorigenesis and progression of OS. Thus, circ_0001105 holds huge promise as a promising therapeutic target for OS.
Previous studies have reported that circRNAs may regulate gene expression by serving as miRNA sponges.27 To further explore the molecular mechanism of circ_0001105 involving in the progression of OS, we assessed whether that circ_0001105 could bind to miR-766. Jia et al found that miR-766 expression was increased in colorectal cancer and could function as an oncogene.28 Similarly, Li et al demonstrated that miR-766 promoted cell proliferation of human colorectal cancer by inhibiting the expression of SOX6.29 However, the expression status and biological function of miR-766 in OS are largely unknown. Similar to the results observed in colorectal cancer, we found miR-766 was significantly upregulated in OS tissues and cell lines. Functional experiments demonstrated that miR-766 mimics partly reversed the prompt-proliferation and invasion effect caused by hsa_circ_0001105 overexpression. These findings suggest that miR-766 may function as an oncogene in OS. Opposite findings were observed in lung cancer and hepatocellular carcinoma, which indicates the tumor-suppressor role of miR-766.30–32 in cancer vary with the cell type or conditions. Thus, the function role of miR-766 needs to be further investigated to draw precision conclusions.
miRNAs exert their biological functions through post-transcriptional regulating gene expression. Our findings show that YTH N6-Methyladenosine RNA Binding Protein 2 (YTHDF2) is a functional target of miR-766, while miR-766 inhibits the progression of OS by targeting YTHDF2. YTHDF2 is an m6A reader protein that promotes mRNA degradation by targeting m6A-mRNAs to the processing bodies in the cytoplasm.33 It has been reported that YTHDF2 plays dual roles in cancer. YTHDF2 could suppress cell proliferation in acute myeloid leukemia and hepatocellular carcinoma.34,35 Opposite results were observed in gastric cancer, prostate cancer and lung cancer.36–38 Here, we report that YTHDF2 is downregulated in OS tissues and inhibited OS cell growth and invasion viability. Besides, miR-766 overexpression or hsa_circ_0001105 silencing counteract the effects of YTHDF2. Moreover, the expression level of YTHDF2 was significantly up-regulated, whereas miR-766 was distinctly decreased after hsa_circ_0001105 overexpression. Our findings suggest that hsa_circ_0001105 may act as a ceRNA (competing endogenous RNA) of miR-766 to relieve the repressive effect of miR-766 on its target YTHDF2.
Despite these results, there are few limitations and shortcomings in the current study. First, one gene may be targeted and regulated by multiple molecules; other circRNA/miRNAs might also contribute to the osteosarcoma progression through regulating YTHDF2. Secondly, YTHDF2 is an m6A regulator. Whether YTHDF2 affects the progression of osteosarcoma through an m6A modification pattern remains to be clarified.
Taken together, this study identified the decreased expression of circ_0001105 in OS tissues. A functional study revealed that circ_0001105 suppresses the progression of OS by modulating the circ_0001105/miR-766/YTHDF2 axis. Although the functional roles and molecular mechanisms of circRNAs remain largely unclear, our study underscores the role and therapeutic potential of the circ_0001105/miR-766/YTHDF2 in OS.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Ca a Cancer J Clin. 2018;60(5):277–300.
2. Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3.
3. Bacci G, Longhi A, Versari M, Mercuri M, Briccoli A, Picci P. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer. 2010;106(5):1154–1161. doi:10.1002/(ISSN)1097-0142
4. Beermann J, Piccoli M-T, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
5. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, NY). 2013;19(2):141–157. doi:10.1261/rna.035667.112
6. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20. doi:10.1038/s41580-018-0059-1
7. Wang D, Yang S, Wang H, et al. The progress of circular RNAs in various tumors. Am J Transl Res. 2018;10(6):1571–1582.
8. Rong J, Wang Q, Zhang Y, et al. Circ-DLG1 promotes the proliferation of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11(undefined):6723–6730. doi:10.2147/OTT
9. Cao S, Chen G, Yan L, Li L, Huang X. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11(undefined):7385–7394. doi:10.2147/OTT
10. Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10(undefined):5783–5794. doi:10.2147/OTT
11. Zhang L, Ding F. Hsa_circ_0008945 promoted breast cancer progression by targeting miR-338-3p. Onco Targets Ther. 2019;12(undefined):6577–6589. doi:10.2147/OTT
12. Wang Y, Li H, Lu H, Qin Y. Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. Onco Targets Ther. 2019;12(undefined):7055–7065. doi:10.2147/OTT
13. Liu C, Zhang Z, Qi D. Circular RNA hsa_circ_0023404 promotes proliferation, migration and invasion in non-small cell lung cancer by regulating miR-217/ZEB1 axis. Onco Targets Ther. 2019;12(undefined):6181–6189. doi:10.2147/OTT
14. Qiao G-L, Chen L, Jiang W-H, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther. 2019;12(undefined):5849–5860. doi:10.2147/OTT
15. Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109(undefined):226–232. doi:10.1016/j.biopha.2018.10.072
16. Zheng S, Qian Z, Jiang F, et al. CircRNA LRP6 promotes the development of osteosarcoma negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11(7):4126–4138.
17. Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33. doi:10.1186/s12943-019-0947-9
18. Liu X, Liu L, Dong Z, et al. Expression patterns and prognostic value of mA-related genes in colorectal cancer. Am J Transl Res. 2019;11(7):3972–3991.
19. Chen J, Yu Y, Chen X, et al. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018;51(11):e12510. doi:10.1111/cpr.2018.51.issue-6
20. Bao J, Yu Y, Chen J, et al. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9(10):1045. doi:10.1038/s41419-018-1020-0
21. Zhang J, He Y, Yu Y, et al. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018;7(7):3351–62.
22. Wang Z, Tang J, Wang Y, et al. Circular RNA hsa_circ_009755 downregulation correlates with clinicopathology in oral squamous cell carcinoma. Onco Targets Ther. 2019;12(undefined):4025–4031. doi:10.2147/OTT
23. Shi Y, Guo Z, Fang N, et al. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;117(undefined):109151. doi:10.1016/j.biopha.2019.109151
24. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi:10.1186/s12943-019-1007-1
25. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi:10.7150/jca.24619
26. Jin J, Chen A, Qiu W, et al. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cell Biochem. 2019;120(9):15678–15687. doi:10.1002/jcb.v120.9
27. Zhao X, Cai Y, Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci. 2019;20(16):undefined.
28. Jia B, Xia L, Cao F. The role of miR-766-5p in cell migration and invasion in colorectal cancer. Exp Ther Med. 2018;15(3):2569–2574. doi:10.3892/etm.2018.5716
29. Li Y-C, Li C-F, Chen L-B, et al. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015;8(undefined):2981–2988. doi:10.2147/OTT.S89459
30. Bai Y, Zhang G, Cheng R, Yang R, Chu H. CASC15 contributes to proliferation and invasion through regulating miR-766-5p/KLK12 axis in lung cancer. Cell Cycle (Georgetown, Tex). 2019;18(18):2323–2331. doi:10.1080/15384101.2019.1646562
31. Liu L, Qi X, Gui Y, Huo H, Yang X, Yang L. Overexpression of circ_0021093 circular RNA forecasts an unfavorable prognosis and facilitates cell progression by targeting the miR-766-3p/MTA3 pathway in hepatocellular carcinoma. Gene. 2019;714(undefined):143992. doi:10.1016/j.gene.2019.143992
32. You Y, Que K, Zhou Y, et al. MicroRNA-766-3p inhibits tumour progression by targeting Wnt3a in hepatocellular carcinoma. Mol Cells. 2018;41(9):830–841. doi:10.14348/molcells.2018.0181
33. Chen X-Y, Zhang J, Zhu J-S. The role of mA RNA methylation in human cancer. Mol Cancer. 2019;18(1):103. doi:10.1186/s12943-019-1033-z
34. Paris J, Morgan M, Campos J, et al. Targeting the RNA mA reader YTHDF2 selectively compromises cancer stem cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25(1):137–148.e136. doi:10.1016/j.stem.2019.03.021
35. Zhong L, Liao D, Zhang M, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442(undefined):252–261. doi:10.1016/j.canlet.2018.11.006
36. Sheng H, Li Z, Su S, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2019.
37. Li J, Meng S, Xu M, et al. Downregulation of N-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N-methyladenosine levels. Oncotarget. 2018;9(3):3752–3764. doi:10.18632/oncotarget.23365
38. Zhang J, Pi J, Liu Y, Yu J, Feng T. Knockdown of YTH N-methyladenosine RNA binding protein 2 (YTHDF2) inhibits proliferation and promotes apoptosis in MGC-803 gastric cancer cells. Xi bao yu Fen Zi Mian Yi Xue Za Zhi. 2017;33(12):1628–1634.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.