Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA 0060745, a Novel circRNA, Promotes Colorectal Cancer Cell Proliferation and Metastasis Through miR-4736 Sponging
Authors Wang X, Ren Y, Ma S, Wang S
Received 30 November 2019
Accepted for publication 13 January 2020
Published 5 March 2020 Volume 2020:13 Pages 1941—1951
DOI https://doi.org/10.2147/OTT.S240642
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Xuebing Wang, Yanyi Ren, Siyao Ma, Shenyu Wang
Integrated TCM and Western Medicine Department, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, People’s Republic of China
Correspondence: Shenyu Wang
Integrated TCM and Western Medicine Department, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, No. 44 Xiaoheyan Road, Dadong District, Shenyang, Liaoning 110042, People’s Republic of China
Tel/Fax +86-24-31916382
Email [email protected]
Purpose: Recent studies have shown that noncoding RNAs (ncRNAs) play essential roles in the development of a number of cancers. Circular RNAs (circRNAs) have been shown to contribute to the progression of colorectal cancer (CRC).
Methods: In this study, the expression levels of circular RNA 0060745 (circ_0060745), and microRNA 4736 (miR-4736) were measured using qRT-PCR. Kaplan–Meier survival analysis and receiver operating characteristic (ROC) analysis were used to evaluate the diagnostic value of circ_0060745. Transwell assay and cell counting kit-8 (CCK8) assay were used to determine the metastatic and proliferative capacity of CRC cells. The expression of chromosome segregation one like (CSE1L) was measured using Western blotting and immunohistochemistry (IHC). In addition, RNA pull-down assay and luciferase assay were performed to verify the targeted binding between miR-473,6 and circ_0060745, and between as miR-4736 and CSE1L.
Results: We showed that circ_0060745 was upregulated in CRC, and was associated with unfavorable clinicopathological characteristics. We also showed that circ_0060745 acted as an oncogene and promoted CRC cell proliferation and metastasis. Circ_0060745 was primarily located in the cytoplasm. Furthermore, miR-4736 was downregulated in CRC, was a downstream target of circ_0060745, and mediated proliferation and metastasis. We showed that circ_0060745 sequestered miR-4736, which resulted in CRC cell proliferation and metastasis. Finally, we showed that CSE1L, a downstream target of miR-4736, was upregulated in CRC and mediated suppression of proliferation and metastasis in CRC.
Conclusion: The results of this study showed that circ_0060745 promoted CRC cell proliferation and metastasis via modulation of miR-4736/CSE1L signaling. The Circ_0060745/miR-4736/CSE1L axis might be a novel target for the treatment of CRC.
Keywords: circ_0060745, CSE1L, miR-4736, proliferation/metastasis, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common malignant disease, and the fourth most frequent cause of cancer-related death worldwide.1 Approximately 25% of patients with CRC present with clinically detectable liver metastases at initial diagnosis.2 Furthermore, 50% of patients with CRC develop colorectal liver metastasis (CLM) during disease progression.3 The natural history of metastatic colorectal cancer (mCRC) is different. However, because of the non-adaptation of surgical treatment, the prognosis of untreated CLM is extremely unfavorable, with median survival rates of less than eight months.4 Therefore, characterization of the mechanisms of CRC might allow for the development of targeted biological therapeutic strategies.
Circular RNAs (circRNAs), a class of noncoding RNAs, are stable, highly conserved, covalently closed RNA transcripts generated by back-splicing of a single pre-mRNA.5 Studies have shown that circRNAs are associated with a number of human diseases, particularly malignancies.6–9 A number of studies focused on circRNAs and CRC have been gradually increasing. Li found that the circRNA CBL.11 suppressed cell proliferation via modulation of the miR-6778-5p/YWHAE axis in CRC.10 Zeng reported that circHIPK3 was significantly upregulated in CRC tissues and cell lines and that circHIPK3 promoted colorectal cancer growth and metastasis by sponging miR-7.11 Chen used circRNA expression profiling to show that 6264 circRNAs were upregulated (including circ_0060745), and 3981 circRNAs were downregulated, in 4 CRC tissue samples compared with 4 paired adjacent tissue samples.12 In this study, we evaluated the role of circ_0060745 in CRC. The results showed that circ_0060745 was upregulated in CRC, and correlated with poor prognosis. Furthermore, we showed that circ_0060745 functioned as an oncogene in CRC.
Circular RNAs are often located in the cytoplasm. Therefore, they can act as sponges of miRNAs to regulate the expression of miRNAs and their target genes.13,14 Circular RNAs have been extensively reported to function as competing endogenous RNAs (ceRNAs) in many malignancies. For example, Su reported that circRNA 0001649 suppressed proliferation and migration of HepG2 and SMMC-7721 hepatocellular carcinoma cells via sponging of miR-127-5p, miR-612, and miR-4688.15 Zhang found that the circRNA circNRIP1 was highly expressed in gastric cancer using RNA-seq analysis, and circNRIP1 promoted gastric cancer progression via regulation of the AKT1/mTOR pathway by sponging microRNA-149-5p.16 In our present study, we showed that circ_0060745 acted as a ceRNA to sequester microRNA-4736 (miR-4736), and promoted chromosome segregation 1-like (CSE1L)-mediated proliferation and metastasis of CRC cells.
Materials and Methods
Patients and Tissue Samples
In this study, we collected 28 paired CRC tissue specimens and paratumor specimens from patients diagnosed with CRC at the Cancer Hospital of China Medical University/Liaoning Cancer Hospital & Institute (Shenyang, China). Three pairs of tissue specimens were used for immunohistochemistry (IHC) analysis. Each of the 28 pairs of tissue specimens was evaluated for expression of circ_0060745, miR-4736, and CSE1L using qRT-PCR.
Sixty formalin-fixed paraffin-embedded (FFPE) CRC specimens were evaluated for expression of circ_0060745. The patients included in this study were seen between March 2011 and October 2017. All patients in this study provided written informed consent. The Institute of Research Medical Ethics Committee of Liaoning Cancer Hospital & Institute granted approval for this study. Each of the included patients was clinically diagnosed based on definitive pathological diagnosis, and clinical stages were determined according to the tumor, node, and metastasis (TNM) classification of the International Union Against Cancer (UICC).
Cell Culture
A human colon epithelial cell line (NCM460) and four human CRC cell lines (HT29, LOVO, PKO, and SW480) were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in RPMI1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA), 100 IU/mL penicillin and 100 mg/mL streptomycin (Abcam, Cambridge, MA, UK). All cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Treatment with RNase R
This procedure was performed as previously described.17 Two milligrams of RNA was incubated for 30 min at 37°C with or without 5 U/μg of RNase R (Epicentre Technologies, Madison, WI, USA), purified using an RNeasy MinElute Cleaning Kit (Qiagen, Valencia, CA, USA), then analyzed using RT-PCR.
Actinomycin D Assay
This procedure was performed as previously reported.17 We exposed HT29 cells to 2 μg/mL of actinomycin D (Sigma, St. Louis, MO, USA) at the indicated time points. The cells were harvested, and total RNA was extracted. The expression levels of circ_0060745 and linear CSE1L mRNA were analyzed using qRT-PCR.
Extraction of RNA and Quantitative Real-Time PCR (qRT-PCR)
These procedures were performed as previously described.17 Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). To detect circ_0060745, total RNA was treated with RNase R (Epicentre Technologies) to improve the purity of circ_0060745. MiScript Reverse Transcription Kit (Qiagen) was used for reverse transcription with or without 500 ng of total RNA treated with RNase R, and qRT-PCR was performed using diluted cDNA, special primers, and SYBR Green Mix (Thermo Fisher Scientific). Beta-actin and small nuclear U6 were used as internal controls for circ_0060745, CSE1L, and miRNAs (miR-4736, miR-577, and miR-126). Primer sequences are listed in Table 1.
 |
Table 1 Primer and oligonucleotide sequences used in the present research |
Western Blot Analysis
Western blot analysis was performed as previously described.18 Total proteins were extracted using RIPA lysis buffer (Sigma, St. Louis, MO, USA). Twenty-microgram samples were separated using 10% SDS-PAGE and transferred to PVDF membranes (Merck Millipore, Billerica, MA, USA). After blocking for nonspecific binding, the membranes were incubated with primary antibodies (Abcam, 1:1000 for CSE1L and 1:500 for GAPDH) at 4°C overnight. The membranes were then incubated with a secondary antibody (Abcam, 1:2000) at 24°C for 1 h. Signals of targeted proteins were detected using an ECL detection system.
Plasmid and Oligonucleotide Transfection
Specific circ_0060745 small interfering RNAs (sicirc-1 and sicirc-2) and negative control siRNAs (siNCs), specific CSE1L siRNA (siCSE1L) and corresponding scrambled control siRNA (siSCR), and specific overexpression plasmids loaded with circ_0060745 (oecirc) and CSE1L (oeSCE1L) were purchased from GenePharma (Shanghai, China). MicroRNA 4736 mimic and negative control (NC mimic), and miR-4736 inhibitors and negative control (NC inhibitor) were purchased from RiboBio Co, Ltd (Ribobio, Guangzhou, China) for upregulation and downregulation of miR-4736. All oligonucleotides and plasmids were transfected into LOVO and HT29 cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Cell Counting Kit-8 (CCK8) Assay
The procedure was performed as previously described.19 In brief, HT29 and LOVO cells were seeded in 96-well plates (2 × 103 cells/well), supplemented with 200 µL of culture medium, and incubated at 37°C in a 5% CO2 atmosphere. At days 1, 2, 3, 4, and 5, 20 μL of CCK8 (Keygen, Nanjing, China) solution was added to each well, and the cells were incubated for 2 h. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Transwell Assay
This procedure was performed as previously reported.20 Briefly, HT29 and LOVO cells (with incubation density of 5 × 104) were incubated on upper chambers (Corning, Corning, NY, USA) coated with Matrigel (BD Biosciences) for migration, or uncoated for invasion, in serum-free medium. Medium supplemented with 10% FBS was placed in the lower chambers. After incubation for 24 h, the CRC cells that had migrated or invaded were fixed, stained, and counted using an inverted microscope (Olympus, Tokyo, Japan).
Dual-Luciferase Reporter Assay
This procedure was performed as previously described.21 For the luciferase reporter assay, pmirGLO dual-luciferase vectors (GenePharma, Shanghai, China) were used to construct dual-luciferase reporter plasmids. Then, HT29 or LOVO cells were co-transfected with plasmids and microRNA, and luciferase activity was measured using the dual-luciferase reporter kit (Promega, Madison, WI, USA). Relative firefly luciferase activity was normalized to Renilla luciferase activity.
RNA Pull-Down Assay
The procedure was performed as previously described.22 Wild-type and mutant circ_0060745 transcripts were biotin-labeled using a Biotin RNA Labeling Mix (Roche, Basel, Switzerland) and T7 RNA polymerase (Roche), treated with RNase-free DNase I (Roche) and purified using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). One milligram of whole-cell lysates from HT29 cells or LOVO cells was incubated with 3 μg of purified biotinylated transcripts for 1 h at 25°C. Complexes were isolated using streptavidin agarose beads (Invitrogen). Any RNA present in the pull-down material was detected using real-time PCR analysis to determine the enrichment of miRNAs.
Immunohistochemistry (IHC) Assay
The procedure was performed as previously described.23 Colorectal cancer specimens were fixed in paraformaldehyde fixation, decalcified, dehydrated, paraffin-embedded, then sliced. Sections were incubated with primary antibodies (anti-CSE1L, Abcam, 1:1000) at 4 °C overnight. Then, the slices were incubated with a secondary antibody (Abcam) at 37 °C for 30 min. Slices were then incubated with streptavidin horseradish peroxidase for an additional 30 min (LSAB kit; Dako, Glostrup, Denmark) and stained with 3,3-diaminobenzidine (DAB). Slices were counterstained with hematoxylin, dehydrated, and mounted.
Statistical Analysis
Data were collected from three independent experiments for each analysis, and results were expressed as means ± SD. Data analysis was performed using GraphPad Prism V5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software and SPSS 19.0 statistical software (IBM, Armonk, New York, USA). Correlations between circ_0060745 and clinicopathological features of patients with CRC were analyzed using Pearson’s chi-squared test. Survival analysis was performed using the log-rank test. Student’s t-test was used to determine differences between the two groups. One-way analysis of variance was used to determine differences among three or more sets of data. Differences were considered significant when *p < 0.05.
Results
Circular RNA 0060745 Was Upregulated in Patients with CRC and Correlated with Poor Prognosis
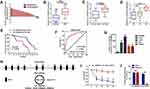
We determined the expression of circ_0060745 in 28 CRC tissue specimens and paired paratumor tissue specimens. As shown in Figure 1A and B, circ_0060745 was upregulated in most (25/28, 89.29%) CRC tissue specimens (p < 0.0001). Furthermore, circ_0060745 was upregulated to a greater degree in CRC tissue specimens from patients with liver (Figure 1C) and lymph-node metastases (Figure 1D). We also measured circ_0060745 expression in 60 paraffin-embedded CRC tissue samples, and classified patients into high and low circ_0060745 groups according to the median value. As shown in Figure 1E and Table 2, high circ_0060745 levels significantly correlated with shorter survival time (p = 0.0002), advanced clinical stage (p = 0.038), nodal (N) classification (p = 0.009), metastasis (M) classification (p = 0.018), and liver metastasis (p = 0.037). Furthermore, an AUC value of 0.8442 (95% confidence interval: 0.7737–0.9147) obtained from ROC curve analysis indicated that circ_0060745 may be a biomarker of CRC (Figure 1F). We then measured the expression of circ_0060745 in CRC cell lines. Quantitative RT-PCR showed that circ_0060745 was upregulated in four CRC cell lines (HT29, LOVO, RKO, and SW480) compared with a normal human colon epithelial cell line (NCM460) (Figure 1G; p < 0.001). Comparison of circ_0060745 with its precursor gene CSE1L showed that circ_0060745 was derived from exon 9 to exon 10 of linear CSE1L (spliced mature full length was 412 bp) (Figure 1H and Supplementary Figure 1A and B). Furthermore, we evaluated the stability of circ_0060745. The transcription inhibitor actinomycin D was added to NCM460 cells, and the expression of circ_0060745 and linear CSE1L mRNA was detected across a range of time points using qRT-PCR. As shown in Figure 1I, circ_0060745 had a half-life of more than 24 hrs, whereas linear CSE1L mRNA had a half-life of fewer than 4 hrs (p < 0.01). We then used RNase R assay to determine the stability of circ_0060745 further. As shown in Figure 1J, the expression of circ_0060745 did not change in response to RNase R (p > 0.05), but the expression of linear CSE1L mRNA decreased substantially following treatment with RNase R (p < 0.01).
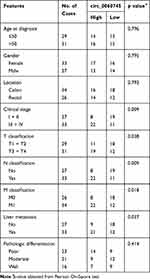 |
Table 2 Correlation of Circ_0060745 Expression and Clinicopathological Features in CRC |
Circular RNA 0060745 Promoted Proliferation and Metastasis of HT29 and LOVO Cells
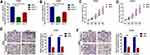
We performed loss of function experiments to determine the role of circ_0060745 in CRC at the cellular level. Specific circ_0060745 siRNAs were used to knock down circ_0060745 in HT29 and LOVO cells. Quantitative RT-PCR results showed a significant reduction in circ_0060745 expression, particularly in response to sicirc-1 (Figure 2A and B). Furthermore, CCK8 assay was used to evaluate the proliferation of HT29 and LOVO cells. As is shown in Figure 2C and D, downregulation of circ_0060745 inhibited HT29 and LOVO cell proliferation (p < 0.01). In addition, the metastatic capacities of HT29 (Figure 2E) and LOVO (Figure 2F) cells were determined using the transwell assay. Knockdown of circ_0060745 suppressed CRC cell migration and invasion (p < 0.01). These findings indicated that circ_0060745 promoted proliferation and metastasis in HT29 and LOVO cells.
MicroRNA 4736 Acted as a Tumor Suppressor in HT29 and LOVO Cells
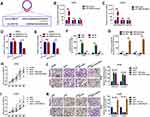
Recent studies indicated that circRNAs function as ceRNAs to sequester multiple miRNAs. We hypothesized that circ_0060745 functioned via a similar mechanism in CRC. Circ_0060745 was mainly located in the cytoplasm (Figure 3A and B), which indicated that circ_0060745 likely acted as an miRNA sponge in CRC. We used online prediction software CircBank, Targetscan, and miRWalk, which resulted in the identification of 6 miRNAs (miR-7-1-3p, miR-1290, miR-219b-3p, miR-4736, miR-6808-5p, and miR-6893-5p) that might be targeted by circ_0060745 (Figure 3C). Analysis of the GEO dataset GSE12218224 resulted in the selection of miR-4736 (probe ID: hsa-miR-4736_st) for further study, as the expression of this microRNA was downregulated in 15 CRC samples compared to that in 10 paratumor samples (Figure 3D and E). Upregulation of miR-4736 suppressed proliferation and metastasis of LOVO and HT29 cells (Figure 3F and G), and vice versa (Figure 3H). These results indicated that miR-4736 acted as a tumor suppressor in CRC cells.
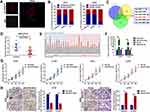 |
Figure 3 MiR-4736 suppressed the proliferation of HT29 and LOVO cells. Notes: (A) Circular RNA 0060745 was primarily located at the cytoplasm, as determined using FISH assay. (B) Relative expression of circ_0060745 in the nuclear and cytoplasmic fractions of HT29 and LOVO cells was determined using qRT-PCR. (C) CircBank (http://www.circbank.cn/), Targetscan (http://www.targetscan.org/vert_71/), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) were used to filter potential miRNAs that might have interacted with circ_0060745. (D) MicroRNA 4736 was expressed at low levels in CRC, as determined by analysis of the GEO dataset GSE122182 (p = 0.0198). (E) The expression of miR-4736 in GSE122182. Probe ID for hsa-miR-4736_st. (F) The expression levels of miR-4736 in HT29 and LOVO cells following transfection with a miR-4736 mimic and miR-4736 inhibitors (miR-4736 inh) were determined using qRT-PCR. **p < 0.01 and ***p < 0.001. (G) Transfection with miR-4736 mimics suppressed cell proliferation, and transfection of miR-4736 inhibitors promoted cell proliferation, in HT29 and LOVO cells. *p < 0.05 and **p < 0.01. (H) The metastatic capacity of HT29 and LOVO cells was evaluated using the transwell chamber migration and invasion assay. **p < 0.01. All data are presented as mean ± SD from three independent experiments. |
Circular RNA 0060745 Promoted Proliferation and Metastasis via Sequestration of miR-4736 in HT29 and LOVO Cells
We evaluated the relationship between circ_0060745 and miR-4736. Circular RNA 0060745 contains eight base pairs of binding sites for miR-4736 (Figure 4A). We performed RNA pull-down assay to evaluate targeted binding between circ_0060745 and miR-4736. A biotinylated circ_0060745 probe was designed and incubated with lysates from HT29 and LOVO cells. Pulled-down miR-4736 was measured using qRT-PCR by using of miR-577 and miR-126 (two previously identified metastasis-related miRNAs in CRC19,25,26) as corresponding control. As shown in Figure 4B and C, miR-4736 was pulled down by circ_0060745.
Luciferase assay was used to confirm the specific binding sites between miR-4736 and circ_0060745. Co-transfection with a luciferase reporter plasmid containing circ_0060745 sequences with wild-type miR-4736 binding sites and miR-4736 mimic resulted in significantly decreased luciferase activity (Figure 4D and E). Mutation of the binding sites for miR-4736 resulted in increased luciferase activity. Furthermore, we showed that circ_0060745 affected miR-4736 with a reciprocal suppression manner (Figure 4F and G). Finally, we evaluated the role of miR-4736 in circ_0060745-induced proliferation and metastasis. As shown in Figure 4H–K, circ_0060745 promoted HT29 and LOVO cell proliferation, and this effect was blocked by miR-4736 mimic. In contrast, knockdown of circ_0060745 suppressed HT29 and LOVO cell proliferation, and this effect was attenuated by inhibition of miR-4736. These findings indicated that circ_0060745 promoted proliferation and metastasis via sequestration of miR-4736.
MicroRNA 4736 Suppressed Proliferation and Metastasis via Targeted Inhibition of CSE1L in HT29 and LOVO Cells
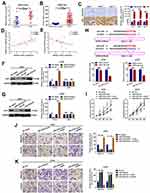
Circular RNAs have been reported to affect the function of their genes of origin via sponging of miRNAs.21,27,28 We showed that circ_0060745 was derived from exon 9 to exon 10 of linear CSE1L and that circ_0060745 sequestered miR-4736. Therefore, we evaluated whether CSE1L was a downstream target gene of the circ_0060745/miR-4736 axis. Analysis of the GEO datasets GSE110224 (probe ID: 201112_s_at) and GSE37364 (probe ID: 201112_at, either) showed that CSE1L was upregulated in CRC (Figure 5A and B and Supplementary Figure 2A and B). Furthermore, we obtained a similar from the online cancer OMICS data analysis software UALCAN (Supplementary Figure 2C and D).29 Immunohistochemistry results showed that CSE1L was upregulated in CRC (Figure 5C). In addition, the expression of CSE1L was positively correlated with circ_0060745, but negatively correlated with miR-4736 (Figure 5D and E). Furthermore, the expression of miR-4736 inversely correlated with CSE1L protein expression, but not CSE1L mRNA expression (Figure 5F and G). MicroRNAs have been shown to regulate downstream genes via directly binding to 3ʹ untranslated regions (3ʹUTR). Luciferase assay showed that miR-4736 bound directly to the 3ʹUTR of CSE1L (position 3215–3221) (Figure 5H). In addition, miR-4736 mimics suppressed proliferation, and this effect was reversed by the upregulation of CSE1L (oeCSE1L) in HT29 and LOVO cells. In contrast, miR-4736 inhibitors induced HT29 and LOVO cell proliferation, and this effect was reversed by CSE1L suppression (Figure 5I). Finally, transwell assay results showed that miR-4736 suppressed HT29 and LOVO cell migration and invasion via CSE1L (Figure 5J and K). These findings indicated that circ_0060745 promoted CRC cell proliferation and metastasis through modulation of the miR-4736/CSE1L axis.
Discussion
Noncoding RNAs contribute to the progression of cancer through the regulation of a variety of molecules. Long noncoding RNAs, circRNAs, and miRNAs have been associated with a number of malignant tumors, including CRC.30–33 The functions of circRNAs include miRNA sponging, regulation of alternative splicing, and modulation of the expression of precursor genes.14,34,35 Recently, the role of circRNAs in CRC has received increased attention. Chen S reported that circ_0060745 was upregulated 2.027748-fold in CRC compared with that in paratumor tissue.12 In the present study, we showed that circ_0060745 was upregulated in most (25/28, 89.29%) CRC tissue specimens. Further clinicopathological evaluation showed that upregulated circ_0060745 was closely correlated with liver metastasis, lymph node metastasis, and shorter survival time in patients with CRC. These findings indicated that circ_0060745 might serve as an oncogene in CRC. Loss of function analyses with CCK8 and transwell assays showed that circ_0060745 promoted CRC proliferation and metastasis.
Circular RNAs are produced by pre-mRNA back-splicing of exons, which results in the exclusion of a circular RNA molecule with a 3ʹ,5ʹ‑phosphodiester bond at the junction site.36 Circular RNAs can modulate gene expression through modulation of transcription, protein binding, sponging of miRNAs, and protein translation.17,37,38 In this study, we showed that circ_0060745 was derived from exon 9 to exon 10 of linear CSE1L. Previous studies have shown that circRNAs are primarily located in the cytoplasm, and function as competitive endogenous RNAs and modulators of miRNA activity by competing for miRNA-binding sites.14,35,39 MicroRNA 577 and miR-126 have been shown to exhibit tumor suppression activity in CRC and was involved in CRC proliferation and metastasis.19,40-42 We found that circ_0060745 was primarily located in the cytoplasm of CRC cells. Therefore, we hypothesized that circ_0060745 acted as an miRNA sponge. Online prediction and bioinformatics analysis resulted in the identification of miR-4736 for further study. Wang reported that miR-4736 was significantly over-expressed in ALK-negative anaplastic large cell lymphoma and was implicated in the pathogenesis of this form of lymphoma.43 We found that miR-4736 acted as a tumor suppressor, and upregulation of miR-4736 inhibited proliferation and metastasis of CRC cells. To further determine the relationship between miR-4736 and circ_0060745, we performed an RNA pull-down assay and luciferase assay. The results showed that circ_0060745 was a target of miR-4736. Furthermore, circ_0060745 promoted CRC cell proliferation and metastasis via miR-4736 sequestration.
Chromosome segregation 1-like (CSE1L) is located at chromosome 20q13.13 and contains 25 exons. Studies have shown that CSE1L is involved in the progression of several cancers.44 Pimiento reported that CSE1L knockdown inhibited cell proliferation and induced apoptosis in CRC cells.45 Li found that CSE1L acted as an oncogene in gastric cancer (GC), and knockdown of CSE1L suppressed GC cell proliferation and metastasis via inhibition of glycoprotein non-metastatic melanoma protein B (GPNMB) expression.46 In this study, analysis of GEO datasets (GSE110224 and GSE37364) and IHC analysis showed that CSE1L was upregulated in CRC. Spearman correlation analysis indicated that CSE1L was positively correlated with circ_0060745 and that CSE1L was inversely correlated with miR-4736. Luciferase assay showed that CSE1L was a direct target of miR-4736, and binding of miR-4736 to CSE1L suppressed proliferation and metastasis of CRC cells.
In summary, this study showed that circ_0060745 may play a role in CRC by promoting oncogenesis, cell proliferation, and metastasis via regulation of CSE1L. This regulation occurred through miR-4736 sponging of SCE1L. These findings demonstrated that circ_0060745 may be a novel target for the treatment of CRC.
Author Contributions
All authors contributed to data analysis, drafting, and revising the article, provided final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this study.
References
1. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet (London, England). 2014;383:1490–1502. doi:10.1016/S0140-6736(13)61649-9
2. De Greef K, Rolfo C, Russo A, et al. Multidisciplinary management of patients with liver metastasis from colorectal cancer. World J Gastroenterol. 2016;22:7215–7225. doi:10.3748/wjg.v22.i32.7215
3. Garden OJ, Rees M, Poston GJ, et al. Guidelines for resection of colorectal cancer liver metastases. Gut. 2006;55(Suppl 3):iii1–iii8. doi:10.1136/gut.2006.098053
4. Bengtsson G, Carlsson G, Hafstrom L, Jonsson PE. Natural history of patients with untreated liver metastases from colorectal cancer. Am J Surg. 1981;141:586–589. doi:10.1016/0002-9610(81)90057-X
5. Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi:10.1016/j.molcel.2015.03.027
6. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi:10.1186/s12943-017-0663-2
7. Li D, Yang Y, Li ZQ, Li LC, Zhu XH. Circular RNAs: from biogenesis and function to diseases. Chin Med J. 2019;132(20):2457.
8. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol cancer. 2019;18:116.
9. Ming L, Jae N, Heumuller AW, Fouani Y, Dimmeler S. Long non-coding RNAs in vascular biology and disease. Mol Cancer. 2019;114:13–22.
10. Li H, Jin X, Liu B, Zhang P, Chen W, Li Q. CircRNA CBL.11 suppresses cell proliferation by sponging miR-6778-5p in colorectal cancer. BMC Cancer. 2019;19:826. doi:10.1186/s12885-019-6017-2
11. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi:10.1038/s41419-018-0454-8
12. Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499–2512. doi:10.3892/or.2018.6372
13. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi:10.1038/ncomms11215
14. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi:10.1038/nature11993
15. Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging. 2019;11:3362–3375. doi:10.18632/aging.101988
16. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20.
17. Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200.
18. Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi:10.1186/s12943-018-0837-6
19. Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50:57. doi:10.1038/s12276-018-0082-5
20. Wang Y, Zhao W, Fu Q. miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol Cell Biochem. 2013;384:105–111. doi:10.1007/s11010-013-1786-4
21. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi:10.1186/s12943-019-0951-0
22. Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi:10.1038/s41467-017-00304-1
23. Wang Y, Zhang Y, Yang T, et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi:10.18632/oncotarget.19727
24. Feng YY, Zeng DZ, Tong YN, et al. Alteration of microRNA-4474/4717 expression and CREB-binding protein in human colorectal cancer tissues infected with fusobacterium nucleatum. PLoS One. 2019;14:e0215088. doi:10.1371/journal.pone.0215088
25. Almeida AL, Bernardes MV, Feitosa MR, et al. Serological under expression of microRNA-21, microRNA-34a and microRNA-126 in colorectal cancer. Acta Cir Bras. 2016;31(Suppl 1):13–18. doi:10.1590/S0102-86502016001300004
26. Zhou Y, Feng X, Liu YL, et al. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013;8:e81203. doi:10.1371/journal.pone.0081203
27. Bai N, Peng E, Xia F, Wang D, Li X, Li X. CircABCC2 regulates hepatocellular cancer progression by decoying MiR-665. J Cancer. 2019;10:3893–3898. doi:10.7150/jca.31362
28. Kong Y, Yang L, Wei W, et al. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics. 2019;11:1163–1176. doi:10.2217/epi-2019-0093
29. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York, NY). 2017;19:649–658. doi:10.1016/j.neo.2017.05.002
30. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi:10.1038/onc.2017.361
31. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi:10.1016/j.canlet.2015.06.003
32. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi:10.1016/j.ccell.2016.03.010
33. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi:10.1016/j.cell.2019.10.017
34. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi:10.1016/j.molcel.2014.08.019
35. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928
36. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi:10.1038/nrm.2015.32
37. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7:4183–4191. doi:10.7150/thno.21299
38. Li X, Liu CX, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–27.e7. doi:10.1016/j.molcel.2017.05.023
39. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi:10.1080/15476286.2015.1020271
40. Sabry D, El-Deek SEM, Maher M, et al. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1alpha-VEGF signaling pathway. Mol Cell Biochem. 2019;454:177–189. doi:10.1007/s11010-018-3462-1
41. Zhang Y, Wang X, Xu B, et al. Epigenetic silencing of miR-126 contributes to tumor invasion and angiogenesis in colorectal cancer. Oncol Rep. 2013;30:1976–1984. doi:10.3892/or.2013.2633
42. Zhou FR, Pan ZP, Shen F, et al. Long noncoding RNA DLX6-AS1 functions as a competing endogenous RNA for miR-577 to promote malignant development of colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23:3742–3748.
43. Wang C, Chen X, Chen X, He Y, Cao L. Expression of microRNA in ALK-negative anaplastic large cell lymphoma and CD30-positive peripheral T cell lymphoma, not otherwise specified. Zhonghua Bing Li Xue Za Zhi. 2015;44:565–570.
44. Jiang MC. CAS (CSE1L) signaling pathway in tumor progression and its potential as a biomarker and target for targeted therapy. Tumour Biol. 2016;37:13077–13090. doi:10.1007/s13277-016-5301-x
45. Pimiento JM, Neill KG, Henderson-Jackson E, et al. Knockdown of CSE1L gene in colorectal cancer reduces tumorigenesis in vitro. Am J Pathol. 2016;186:2761–2768. doi:10.1016/j.ajpath.2016.06.016
46. Li Y, Yuan S, Liu J, et al. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J Cell Physiol. 2019.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.