Back to Journals » International Journal of General Medicine » Volume 13
Circular RNA 0001313 Knockdown Suppresses Non-Small Cell Lung Cancer Cell Proliferation and Invasion via the microRNA-452/HMGB3/ERK/MAPK Axis
Authors Zhang S, Liu J, Yuan T, Liu H, Wan C, Le Y
Received 20 July 2020
Accepted for publication 10 November 2020
Published 10 December 2020 Volume 2020:13 Pages 1495—1507
DOI https://doi.org/10.2147/IJGM.S272996
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Shihao Zhang,* Jiansheng Liu,* Taiwen Yuan, Huiyu Liu, Chengwei Wan, Yonghong Le
Department of Respiratory and Critical Care Medicine, Ganzhou People’s Hospital, Ganzhou 341000, Jiangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yonghong Le
Department of Respiratory and Critical Care Medicine, Ganzhou People’s Hospital, No. 17, Hongqi Avenue, Zhanggong District, Ganzhou 341000, Jiangxi, People’s Republic of China
Tel/Fax +8613879769189
Email [email protected]
Background: Non-small cell lung cancer (NSCLC) seriously endangers human health. Circular RNAs (circRNAs) regulate diverse types of cancers, including NSCLC. This study investigated the possible mechanism of circ0001313 in NSCLC.
Materials and Methods: Circ0001313 expression in NSCLC tissues was measured, and its correlation with clinicopathological features was analyzed. The binding relationships among circ0001313, microRNA (miR)-452 and HMGB3 were tested. The gain and loss of functions were performed to examine NSCLC cell malignant behaviors. After HMGB3 overexpression, ERK/MAPK pathway-related protein levels were detected. Subsequently, the rescue experiment was further performed using an ERK/MAPK pathway inhibitor PD98059.
Results: Abnormally elevated circ0001313 and decreased miR-452 in NSCLC cells were observed. Circ0001313 silencing or miR-452 overexpression significantly reduced NSCLC cell proliferation and invasion. Circ0001313 competitively bound to miR-452 to upregulate HMGB3, thus promoting NSCLC cell growth. HMGB3 overexpression activated the ERK/MAPK pathway to contribute to NSCLC development.
Conclusion: We highlighted that silencing of circ0001313 blunted the ERK/MAPK pathway via the miR-452/HMGB3 axis, thereby inhibiting NSCLC cell proliferation and invasion.
Keywords: non-small cell lung cancer, circular RNA 0001313, microRNA-452, HMGB3, ERK/MAPK pathway, proliferation and invasion
Introduction
Non-small cell lung cancer (NSCLC), accounting for about 85% cases of lung cancer, is featured by late diagnosis, rapid metastasis and high incidence of recurrence, and contributes largely to cancer-related deaths.1,2 NSCLC can be more accurately subdivided into lung adenocarcinoma and lung squamous cell carcinoma.3 NSCLC, as a major consequence of smoking, can lead to cancer anorexic-cachexia, resulting in weight loss, reduced food intake and decreased life quality of patients.4 However, due to its resistance to chemotherapeutic drugs and rapid metastasis characteristics, NSCLC treatment options are limited and the overall survival rates for NSCLC patients remain low.5 Therefore, it is urgent to search for new therapies to improve outcomes in NSCLC.
Circular RNAs (circRNAs), endogenous RNAs with stable structure, have been defined as naturally occurring RNAs with high expression in the eukaryotic transcriptome.6,7 Emerging evidence indicates that circRNAs take part in various cancers, promising to be both diagnostic and therapeutic targets.8 The role of circRNAs in NSCLC progression has also been increasingly appreciated.9 Circ0001313, also known as circCCDC66, is a novel circRNA that has been evidenced to promote cancer development in colon cancer.10 In addition, circ0001313 suppression curtailed cell proliferation by about 65% in A549 cells and by about 40% in H1299 cells, making it a potential biomarker for NSCLC.11 Nevertheless, the underlying mechanism remains largely unknown.
A previous study has suggested that the occurrence and progression of NSCLC is in a close relationship with circRNA-microRNA (miR)-mRNA network.12 miRs, defined as small noncoding RNAs, play a significant part in regulating genic expression and consequently various cellular processes, including cancer development.13 miRs are implicated in the clinical status of NSCLC patients and are advantageous therapeutic agents for NSCLC.14 miR-452 expression was found to be significantly downregulated in colorectal cancer tissues relative to the adjacent noncancerous tissues, and a low level of miR-452 was associated with larger tumor size, deeper invasion depth, and advanced tumor-node-metastasis (TNM) stage.15 More specifically, miR-452 downregulation was significantly associated with tumor differentiation grade and tumor size in NSCLC.16 Interestingly, another circRNA, circKIAA0907 bound to miR-452-5p as a specific sponge for it to participate in the progression of gastric cancer.17
Hence, we speculated that circ0001313 regulates cell behaviors in NSCLC via interacting with miR-452 to form a circRNA-miR-mRNA network. Consequently, we performed a series of histological and molecular experiments to identify the circRNA-miR-mRNA network and to study the underlying molecular machinery, with the purpose to provide some novel therapies against NSCLC progression.
Materials and Methods
Ethics Statement
This study was supervised and approved by the ethics committee of Ganzhou People’s Hospital. All participants signed the informed consent. This study conforms to all relevant ethical norms of research involving human participants.
Sample Collection
NSCLC tissues and adjacent normal tissues from 59 patients undergoing NSCLC resection were collected. All the patient samples were obtained from Ganzhou People’s Hospital. The pathological diagnosis results were obtained according to the histology or biopsy of tumor samples and examined by experienced pathologists. All tissues were stored in liquid nitrogen.
Cell Culture and Transfection
Human lung epithelial BEAS-2B cells without mycoplasma contamination and NSCLC cell lines H1299, A549, NCI-H23 and NCI-H522 were selected. All the above cells were obtained from ATCC (Manassas, Virginia, USA) and cultured in 90% RPMI-1640 medium with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific Inc., Waltham, MA, USA), as well as 100 mg/mL streptomycin and 100 U/mL penicillin in a humidity-saturated incubator with 5% CO2 at 37°C.
Vectors for transfection were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, Guangdong, China). Small interfering RNA (si-RNA) was designed and synthesized by Thermo Fisher (si-circ0001313-#1, F: 5ʹ-CGGCUUACCCUGAGCGGAATT-3ʹ; R: 5ʹ-UUCCGCUCAGGGUAAGCCGTT-3ʹ; si-circ0001313-#2, R: 5ʹ-UUCUCCAGCAGCUCCGCCATT-3ʹ; F: 5ʹ-CGGAGCUGCUGGAGAAGUATT-3ʹ). A549 cells were plated in 6-well plates (1 × 106 cells/well) overnight. The transfection was performed using Lipofectamine 2000 reagent (Invitrogen Inc., Carlsbad, CA, USA). According to different experimental requirements, the transfected cells were collected for subsequent experiments.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol (Sigma-Aldrich, St Louis, MO, USA) after 48-h transfection. RNA enzyme-free ultrapure water was used to dilute 5 μL RNA sample for 20 times. An ultraviolet spectrophotometer was used to measure the optical density (OD) value at 260 nm and 280 nm. RNA concentration and purity were determined with the purity detected by the OD260/OD280 ratio. Reverse transcription was performed on the PCR amplification instrument to synthesize the cDNA template according to the instructions of the kit (Beyotime Biotechnology Co., Ltd., Shanghai, China). The required qPCR primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The primer information is listed in Table 1. The total qPCR reaction volume was 10 μL, including 5 μL 2 × SYBR Premix (Takara Biotechnology Co., Ltd., Dalian, China), 1 μL cDNA template, 0.5 μL forward and reverse primers, and 3 μL double distilled H2O. The conditions for reaction included 30 s at 95°C (pre-denaturation), and then 40 cycles of 30 s at 95°C (denaturation), 20 s and 30 s at 72°C (annealing/extension). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was the internal reference for circ0001313 and high mobility group box 3 (HMGB3), and U6 was the internal reference for miR-452.
 |
Table 1 Primer Sequence for RT-qPCR |
Western Blot (WB)
After 48 h of transfection, cell culture medium was removed. Cells were washed three times with precooled phosphate-buffered saline (PBS) and added with radioimmunoprecipitation assay lysis buffer (MedChemExpress Co., Ltd., Monmouth Junction, NJ, USA) containing 10% protease inhibitor. Cell sample was transferred to a 1.5 mL centrifuge tube and centrifuged at 13,000 g for 10 min, and the supernatant was obtained. The protein concentration was assessed using bicinchoninic acid method. Protein was stored at −20°C. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis for protein separation, proteins were transferred to nitro cellulose membranes using wet transfer method and blocked for 1 h with 5% skim milk. After that, diluted primary rabbit polyclonal antibodies against HMGB3 (27,465-1-AP, Proteintech Group, Inc., Wuhan, Hubei, China), extracellular signal-regulated kinase 1/2 (ERK1/2, ab17942, Abcam Inc., Cambridge, MA, USA), p-ERK1/2 (ab223500, Abcam), p38 mitogen-activated protein kinases (MAPK, 66,234-1-Ig, Proteintech), p-p38 MAPK (ab4822, Abcam) and GAPDH (60,004-1-Ig, Proteintech) were added for incubation at 4°C overnight, followed by 2-h incubation with secondary antibody immunoglobulin G (IgG, ab150077, Abcam) in a shaking table. The membranes were developed using a Bio-Rad gel imaging system (MG8600, Beijing Thmorgan Biotech Ltd., Beijing, China). Quantitation analysis was done using Image-Pro P1us 7.0 software (Media Cybernetics, Singapore).
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 (Bimake, Houston, TX, USA) was conducted for cell proliferation ability detection. Cells were collected after 24 h of transfection and seeded into 96-well plates (3000 cells/well). The OD value was measured at 450 nm with a microplate plate reader.
Transwell Assay
The invasiveness of cells was detected using transwell culture system. The membrane of the apical chamber was coated with Matrigel (BD Biosciences Discovery Labware, Woburn, MA, USA). The chambers were rehydrated with serum-free media for 2 h in a 37°C incubator. The apical chamber was then hydrated with 200 μL cell suspension containing 1 × 105 cells, and 500 μL medium containing 10% FBS was added as chemoattractant to the basolateral chamber. After 24-h incubation at 37°C, the cells invaded were fixed with formaldehyde for 5 min, stained with 10% crystal violet for 10 min and counted under the microscope.
RNA Fluorescence in situ Hybridization (FISH)
Circ0001313 expression in A549 cells was detected by FISH Kits (C10910, RiboBio). Cell slides were placed on the bottom of the 24-well plates. A549 cells growing in logarithmic phase were detached and added to the slides (6 × 104 cells/well). When cells grew to 60–70% confluence, cells were fixed with 4% paraformaldehyde for 10 min, treated with 1 mL precooled permeating solution at 4°C for 5 min, with 200 μL prehybridizing solution for blocking at 37°C for 30 min. At the same time, the hybridizing solution was preheated at 37°C. Under dark conditions, 2.5 μL FISH Probe Mix storage solution (20 μM) was added to the hybridizing solution. Afterwards, prehybridizing solution in each well was discarded, and a proper amount of hybridizing solution containing circ0001313 probe was added for incubation in the dark at 37°C overnight. Next, cells in each well were washed using washing solution I 3 times in the dark at 42°C (5 min each) to reduce the background signal, followed by one wash using washing solution II at 42°C, one wash using washing solution III at 42°C and one wash using 1 × PBS for 5 min. After that, cells were stained for 10 min with 4ʹ,6-diamidino-2-phenylindole staining solution. Finally, cell slides were taken out from wells and fixed with the sealing agent for fluorescence detection. All the above operations were performed under the condition of avoiding light. The specific probe of circ0001313 was synthesized by RiboBio (Dig-5ʹ-CTGGAGCACCCTTTCAAAGGTGTCAGTATG-3ʹ-Dig).
Dual-Luciferase Reporter Gene Assay
The binding sites between circ0001313 and miR-452 were obtained using the bioinformatics prediction website StarBase (http://starbase.sysu.edu.cn/index.php). The fragment sequences containing the binding sites were obtained. Full circ0001313 and HMGB3 3ʹUTR were cloned and amplified, respectively, into pmirGLO luciferase vectors (E1330, Promega Corporation, Madison, WI, USA), named Circ0001313-wild type (WT) and HMGB3-WT. The circ0001313-mutant (MUT) and HMGB3-MUT vectors were constructed, respectively, on the basis of the predicted binding sites. The renilla luciferase-expressing plasmids pRL-TK (E2241, Promega) was used as an internal reference. miR-452 mimic and mimic negative control (NC) were co-transfected with luciferase report vectors into 293T cells, respectively. The fluorescence intensity was detected using a fluorescence detector (Glomax20/20, Promega).
RNA Pull-Down
Cells were transfected with 50 nM biotin-labeled WT-bio-miR-452 and MUT-bio-miR-452, followed by a 10-min incubation in a specific lysis buffer (Ambion, Austin, TX, USA). The lysate was incubated with M-280 streptavidin beads (S3762, Sigma-Aldrich) precoated with RNAse-free BSA and Yeast tRNA (TRNABAK-RO, Sigma-Aldrich). The beads were incubated at 4°C for 3 h, washed twice with precooled lysis buffer, 3 times with low-salt buffer and once with high-salt buffer. The binding RNA was purified using TRIzol and then detected.
RNA Immunoprecipitation (RIP)
Cells were lysed using lysis buffer containing 25 mM Tris-HCl (pH = 7.4), 150 mM NaCl, 0.5% Nonidet P-40, 2 mM ethylene diamine tetraacetic acid, 1 mM NaF and 0.5 mM dithiothreitol plus a mixture of RNasin (Takara) and protease inhibitor (B14001a, Roche Diagnostics, Indianapolis, Indiana, USA). The lysate was subjected to a 30 min-centrifugation at 12,000 g. The supernatant was then incubated with anti-Ago-2 magnetic beads (BMFA-1, Biomarker Technologies Corporation, Beijing, China), with the addition of anti-IgG magnetic beads as the control. After being incubated for 4 h at 4°C, the beads were rinsed 3 times with washing buffer containing 50 mM Tris-HCl, 300 mM NaCl (pH = 7.4), 1 mM MgCl2 and 0.1% NP-40. RNA was extracted from magnetic beads using TRIzol and then detected.
Statistical Analysis
All statistical analysis was conducted using SPSS 18.0 software (IBM Corp., Armonk, NY, USA). The comparison between two groups was analyzed using both upaired and paired t-test. One-way or two-way analysis of variance (ANOVA) was used for analysis of the single-factor and multi-factor comparisons among multiple groups, respectively. All data were expressed as the mean ± standard deviation of at least three independent experiments. For skew distribution data, nonparametric test was used to determine statistical significance. p < 0.05 indicated that the difference was statistically significant.
Results
Circ0001313 is Upregulated in NSCLC and Circ0001313 Downregulation Inhibits NSCLC Cell Proliferation and Invasion
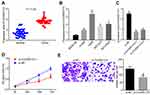
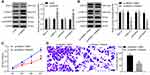
To explore circ0001313 expression in NSCLC, we first collected 59 cases of NSCLC tissues and adjacent normal tissues. The relationship between circ0001313 expression and clinicopathological factors is shown in Table 2. There was no significant correlation between circ0001313 expression and gender, age and lymph node metastasis of NSCLC patients (all p > 0.05), while circ0001313 expression was correlated with tumor diameter, tissue differentiation degree and TNM stage (all p < 0.05). qRT-PCR found that circ0001313 expression in NSCLC tissues was obviously upregulated (Figure 1A). Circ0001313 expression in human lung epithelial BEAS-2B cells without mycoplasma contamination and NSCLC cell lines H1299, A549, NCI-H23 and NCI-H522 was detected using qRT-PCR. Circ0001313 expression was increased in different degrees in NSCLC cell lines relative to that in BEAS-2B cells (Figure 1B). A549 cells with the highest circ0001313 expression were selected for subsequent experiments.
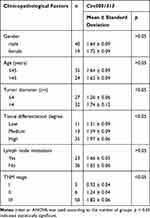 |
Table 2 Relationship Between Circ0001313 Expression and Clinicopathological Characteristics |
To explore the effect of circ0001313 expression on NSCLC cell proliferation and invasion, we cultured A549 cells and transfected cells with si-NC and si-circ0001313. Transfection efficiency was verified using qRT-PCR. si-circ0001313-1 and si-circ0001313-2 were designed, and one with better effect was selected for subsequent experiments (Figure 1C). Cell transfected with si-circ0001313 showed remarkably reduced cell proliferation ability (Figure 1D). Transwell assay demonstrated that cell invasion ability following si-circ0001313 treatment was significantly decreased (Figure 1E). These results indicated that circ0001313 showed promoting effects on NSCLC cell proliferation and invasion.
miR-452 Competitively Binds to Circ0001313
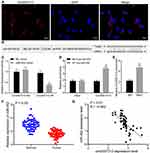
RNA-FISH detected the location of circ0001313 in A549 cells and showed that circ0001313 was mainly located in the cytoplasm (Figure 2A). A binding site between circ0001313 and miR-452 was predicted through Starbase (Figure 2B). Dual-luciferase reporter gene assay verified the interaction between circ0001313 and miR-452. miR-452 overexpression dramatically inhibited the luciferase activity of circ0001313 wt but had little impact on circ0001313 mut (Figure 2C). RNA pull-down showed that compared with MUT-miR-452, WT-miR-452 obviously bound to more circ0001313, indicating that miR-452 and circ0001313 could directly bind to each other (Figure 2D). RIP results showed that compared with IgG, Ago2 remarkably bound more circ0001313 (Figure 2E), indicating that circ0001313 could bind directly to Ago2 protein, namely, miR-452 directly bound to circ0001313. And then, we further detected the expression of miR-452 in tumor and adjacent tissues of 59 patients. The expression of miR-452 in NSCLC tissues was observed to be significantly lower than that in adjacent tissues (Figure 2F). The correlation between miR-452 expression and circ0001313 expression was analyzed and we found a negative correlation (Figure 2G). The above results indicated that miR-452 competitively bound to circ0001313.
Overexpression of miR-452 Inhibits NSCLC Cell Proliferation and Invasion
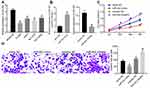
miR-452 expression in BEAS-2B cells and NSCLC cell lines was detected. The results showed that compared with that in BEAS-2B cells, NSCLC cell lines exhibited decreased miR-452 expression in different degrees with the lowest expression in A549 cells (Figure 3A). To explore the influence of miR-452 expression on NSCLC cell growth, we transfected mimic NC, miR-452 mimic, inhibitor NC and miR-452 inhibitor, respectively, into A549 cells, and the transfection efficiency was verified using qRT-PCR (Figure 3B). CCK-8 assay indicated that compared with cells in the mimic NC group, cells in the miR-452 mimic group showed significantly reduced cell proliferation ability; compared with cells in the inhibitor NC group, cells in the miR-452 inhibitor group exhibited obviously increased cell proliferation ability (Figure 3C). Transwell assay observed that the miR-452 mimic group showed weakened cell invasion ability relative to the mimic NC group; the miR-452 inhibitor group exhibited enhanced cell invasion ability compared with the inhibitor NC group (Figure 3D). From all above, miR-452 played an inhibitory role in NSCLC cell proliferation and invasion.
miR-452 Targets HMGB3 to Regulate NSCLC Cell Proliferation and Invasion
To explore the downstream regulatory mechanism of miR-452, the bioinformatics website Starbase was utilized. miR-452 was predicted to directly bind to the 3ʹ-UTR sequence of HMGB3 (Figure 4A). HMGB3, a key cell cycle regulator, is implicated in various cancers with its high expression. Dual-luciferase reporter gene assay verified the targeting relationship between miR-452 and HMGB3, and overexpression of miR-452 significantly repressed the luciferase activity of HMGB3 wt, but rarely changed the luciferase activity of HMGB3 mut (Figure 4B). WB detected HMGB3 protein expression in each group. HMGB3 protein expression decreased notably after miR-452 overexpression and increased obviously after miR-452 silencing (Figure 4C). A549 cells were transfected with mimic NC + overexpressed (oe)-NC, miR-452 mimic + oe-NC, mimic NC + oe-HMGB3 and miR-452 mimic + oe-HMGB3. qRT-PCR and WB verified the expression of miR-452 and HMGB3. Relative to that in the mimic NC + oe-NC group, miR-452 expression was significantly increased, and HMGB3 levels were obviously reduced in the miR-452 mimic + oe-NC group; miR-452 expression in the mimic NC + oe-HMGB3 showed no significant difference, while HMGB3 levels were noticeably increased. Compared with the miR-452 mimic + oe-NC group, the miR-452 mimic + oe-HMGB3 group showed no obvious difference in miR-452 expression, while exhibited clearly increased HMGB3 levels (Figure 4D and E). Compared with those in the mimic NC + oe-NC group, cells in the miR-452 mimic + oe-NC showed remarkably reduced cell proliferation ability, while that of the mimic NC + oe-HMGB3 group was significantly increased. Cell proliferation in the miR-452 mimic + oe-HMGB3 group was clearly increased compared to the miR-452 mimic + oe-NC group (Figure 4F). Relative to cells in the mimic NC + oe-NC group, cells in the miR-452 mimic + oe-NC group showed significantly weakened cell invasion ability, which was notably enhanced in the mimic NC + oe-HMGB3 group. The cell invasion ability of the miR-452 mimic + oe-HMGB group was obviously increased relative to that of the miR-452 mimic + oe-NC group (Figure 4G). Moreover, we further examined the expression of HMGB3 in tumor and adjacent tissues of 59 lung cancer patients. The expression of HMGB3 in NSCLC tissues was significantly higher than in adjacent tissues (Figure 4H). Furthermore, HMGB3 was negatively correlated with miR-452 and positively correlated with circ0001313 (Figure 4I). Subsequently, we detected the expression of miR-452 and HMGB3 in A549 cells with poor expression of circ0001313. The expression of HMGB3 was significantly decreased after knocking down circ0001313, whereas the expression of miR-452 was significantly increased (Figure 4J). In brief, miR-452 suppressed NSCLC cell proliferation and invasion via targeting HMGB3.
Circ0001313/miR-452/HMGB3/ERK/MAPK Axis Regulates NSCLC Cell Growth
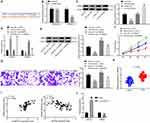
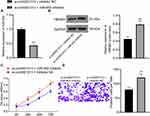
The expression of ERK1/2, p-ERK1/2, p38 MAPK and p-p38 MAPK in the ERK/MAPK pathway was detected after HMGB3 overexpression. p-ERK1/2 and p-p38 MAPK expression was noticeably higher in cells overexpressing HMGB3 (Figure 5A). Therefore, we speculated that HMGB3 might regulate NSCLC cell proliferation and invasion via the ERK/MAPK pathway. Hence, the ERK/MAPK pathway inhibitor PD98059 (50 μmol/L, Selleck Chemicals, Houston, TX, USA) was used. p-ERK1/2 and p-p38 MAPK expression of cells treated with the oe-HMGB3 + PD98059 was obviously reduced relative to that in cells treated with oe-HMGB3 + DMSO (Figure 5B). Compared with those in the oe-HMGB3 + DMSO group, cells in the oe-HMGB3 + PD98059 group showed clearly reduced cell proliferation ability (Figure 5C). Cells in the oe-HMGB3 + PD98059 group exhibited remarkably weakened cell invasion relative to those in the oe-HMGB3 + DMSO group (Figure 5D). In conclusion, circ0001313 targeted miR-452 to upregulate HMGB3 and activate the ERK/MAPK pathway, thus promoting NSCLC cell growth.
miR-452 Inhibitor Abolishes the Inhibitory Effects of Si-Circ0001313 on Proliferation and Invasion of A549 Cells
To further clarify the regulatory relationship between miR-452 and circ0001313, we transfected miR-452 inhibitor into A549 cells with poor expression of circ0001313, and we detected successful transfection by qRT-PCR (Figure 6A). We then further used WB to detect the expression of HMGB3 in cells and found that knocking down the expression of miR-452 resulted in an increase in the protein expression of HMGB3 in cells (Figure 6B). Subsequently, we assayed cell activity by CCK-8 kits. miR-452 inhibitor significantly promoted A549 cell activity in the presence of si-circ0001313 (Figure 6C). Moreover, we found by Transwell assays that knocking down miR-452 significantly restored the number of cells invaded into the Transwell basolateral compartment (Figure 6D).
Discussion
NSCLC represents the most common type of lung cancer and the most frequent cause of mortality in lung cancer patients.5 In China, the high incidence of lung cancer is mainly influenced by tobacco consumption and environmental pollution.18 CircRNAs are members of the noncoding RNAs, and their diverse functions include adsorbing miRNAs by acting as miRNA sponges, regulating transcription, interacting with RNA-binding proteins, as well as translating pseudogenes.19 We highlighted that circ0001313 knockdown regulated the miR-452/HMGB3 axis and impaired the activation of the ERK/MAPK pathway, thereby inhibiting NSCLC cell proliferation and invasion.
CircRNAs are non-coding RNAs that participate in the regulation of various human cancers, including NSCLC.20 Circ0001313 is highly expressed in colon cancer tissues and cell lines, and depletion of circ0001313 repressed cell proliferative ability but induced apoptosis of colon cancer cells.21 Similarly, this study observed the aberrant upregulation of circ0001313 in NSCLC tissues and cell lines. Our data showed that there were significant correlations between circ0001313 expression and tumor diameter, tissue differentiation degree and TNM stage. Moreover, we revealed that downregulated circ0001313 suppressed NSCLC cell proliferation and invasion. Similar to our results, circ0001313 was found to be elevated in NSCLC cells, and circ0001313 knockdown showed a suppressive effect on NSCLC cell growth.22 It has also been demonstrated that circRNAs possess the ability to bind to miRs via acting as molecular sponges.23 hsa-circ0001313 is also found to serve a regulatory part in gastric cancer via targeting miR-618.24 Furthermore, a previous study has shown the interactions between another circRNA and miR-452-5p in hepatocellular carcinoma.25 We identified the competitively binding relationship between circ0001313 and miR-452. In addition, miR-452 was downregulated in NSCLC cells, and overexpression of miR-452 inhibited NSCLC cell proliferation and invasion. miR-452 expression, as a significant predictor in NSCLC patients, decreased dramatically in NSCLC.16 The relatively low expression of miR-452 promoted cell invasive ability and strongly related to the advanced tumor stage in NSCLC.26 In addition, the inhibitory role of miR-452 in proliferation, invasion, and migration of A549 or H460 cells has been previously highlighted.27 Overall, circ0001313 competitively bound to miR-452, thus promoting NSCLC cell proliferation and invasion.
Then, we turned to investigate the downstream mechanism of miR-452. Through the screening of bioinformatics website Starbase and the verification of dual-luciferase reporter gene assays, we identified that miR-452 targeted HMGB3 to inhibit NSCLC cell growth. Previously, HMGB3 overexpression is significantly correlated with tumor grade, tumor size, clinical stage, lymph node metastases, and a poor prognosis of NSCLC patients.28 As for its association with miRNAs, HMGB3 is evidenced to be inversely regulated by miR-452-5p and its knockdown abolished the promotive effects of miR-452-5p inhibitor on growth and metastasis of prostate cancer cells.29 Moreover, HMGB3 serves a regulatory role in NSCLC with its significantly upregulated expression indicating aggravated lymph node metastases and poor prognosis of NSCLC patients.28 HOXA transcript at the distal tip, a lncRNA, has been reported to positively regulate HMGB3 expression by acting as a molecular sponge of miR-615-3p in NSCLC cells.30 More relevantly, two ceRNA networks involving circEPSTI1/miR-145/HMGB3 and circRNA_102179/miR-330-5p/HMGB3 have been documented in NSCLC.31,32 Taken together, circ0001313 promoted NSCLC cell growth via targeting miR-452 to upregulate HMGB3.
Subsequently, we shifted to exploring the downstream possible signaling pathway of HMGB3. The ERK/MAPK pathway has been recently evidenced to closely related to NSCLC cell development.33,34 Inhibited ERK/MAPK pathway is implicated in suppressed NSCLC cell episodes.35 Moreover, previous studies have verified the relationship between HMGB3 and the ERK/MAPK pathway. For example, HMGB3 knockdown remarkably reduced p-ERK 1/2, and HMGB3 contributed to glioblastoma multiforme oncogenesis via the MAPK pathway activation.36 We highlighted that upregulated HMGB3 activated the ERK/MAPK pathway to promote NSCLC cell proliferation and invasion. HMGB3 induced innate immune responses, including MAPK activation.37 Taken together, circ0001313 stimulated NSCLC cell growth via the miR-452/HMGB3 axis and the ERK/MAPK pathway activation.
Conclusion
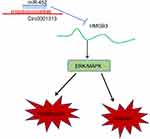
All in all, this study supported that circ0001313 competitively bound to miR-452 to promote HMGB3 expression, thus activating the ERK/MAPK pathway and promoting NSCLC cell growth (Figure 7). Targeting the circ0001313/miR-452/HMGB3 axis might develop as a new therapeutic approach for NSCLC treatment. Although the current study provides a new perspective for the treatment of NSCLC, the clinical application effect needs to be further verified.
Funding
There is no funding to report.
Disclosure
The authors reported no conflicts of interest in this work.
References
1. Peters S, Kerr KM, Stahel R. PD-1 blockade in advanced NSCLC: A focus on pembrolizumab. Cancer Treat Rev. 2018;62:39–49. doi:10.1016/j.ctrv.2017.10.002
2. Zheng H, Zhan Y, Liu S, et al. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J Exp Clin Cancer Res. 2018;37(1):226.
3. Qiu ZW, Bi JH, Gazdar AF, Song K. Genome-wide copy number variation pattern analysis and a classification signature for non-small cell lung cancer. Genes Chromosomes Cancer. 2017;56(7):559–569. doi:10.1002/gcc.22460
4. Currow D, Temel JS, Abernethy A, Milanowski J, Friend J, Fearon KC. ROMANA 3: a Phase 3 safety extension study of anamorelin in advanced non-small-cell lung cancer (NSCLC) patients with cachexia. Ann Oncol. 2017;28(8):1949–1956. doi:10.1093/annonc/mdx192
5. Lobb RJ, van Amerongen R, Wiegmans A, Ham S, Larsen JE, Moller A. Exosomes derived from mesenchymal non-small cell lung cancer cells promote chemoresistance. Int J Cancer. 2017;141(3):614–620. doi:10.1002/ijc.30752
6. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi:10.7150/thno.21299
7. Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi:10.1016/j.prp.2017.02.011
8. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
9. Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889. doi:10.1007/s00432-019-03045-4
10. Wang L, Peng X, Lu X, Wei Q, Chen M, Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol Res Pract. 2019;215(4):689–696. doi:10.1016/j.prp.2018.12.032
11. Hong W, Yu S, Zhuang Y, Zhang Q, Wang J, Gao X. SRCIN1 regulated by circccdc66/miR-211 is upregulated and promotes cell proliferation in non-small-cell lung cancer. Biomed Res Int. 2020;2020:5307641. doi:10.1155/2020/5307641
12. Cai X, Lin L, Zhang Q, Wu W, Su A. Bioinformatics analysis of the circRNA-miRNA-mRNA network for non-small cell lung cancer. J Int Med Res. 2020;48(6):300060520929167. doi:10.1177/0300060520929167
13. Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8(2):61–81.
14. Weidle UH, Birzele F, Nopora A. MicroRNAs as potential targets for therapeutic intervention with metastasis of non-small cell lung cancer. Cancer Genomics Proteomics. 2019;16(2):99–119. doi:10.21873/cgp.20116
15. Tang T, Zhang GC, Li CF, Liu YF, Wang WY. Decreased miR-452 expression in human colorectal cancer and its tumor suppressive function. Genet Mol Res. 2016;15:2. doi:10.4238/gmr.15027730
16. He Z, Xia Y, Liu B, et al. Down-regulation of miR-452 is associated with poor prognosis in the non-small-cell lung cancer. J Thorac Dis. 2016;8(5):894–900. doi:10.21037/jtd.2016.03.51
17. Zhu L, Wang C, Lin S, Zong L. CircKIAA0907 retards cell growth, cell cycle, and autophagy of gastric cancer in vitro and inhibits tumorigenesis in vivo via the miR-452-5p/KAT6B axis. Med Sci Monit. 2020;26:e924160. doi:10.12659/MSM.924160
18. Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer. 2012;131(12):2724–2732. doi:10.1002/ijc.27816
19. Wu J, Qi X, Liu L, et al. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi:10.1016/j.omtn.2019.04.011
20. Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. doi:10.1186/s12943-018-0889-7
21. Tu FL, Guo XQ, Wu HX, et al. Circ-0001313/miRNA-510-5p/AKT2 axis promotes the development and progression of colon cancer. Am J Transl Res. 2020;12(1):281–291.
22. Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019. doi:10.1016/j.biopha.2020.110019
23. Filippenkov IB, Kalinichenko EO, Limborska SA, Dergunova LV. Circular RNAs-one of the enigmas of the brain. Neurogenetics. 2017;18(1):1–6. doi:10.1007/s10048-016-0490-4
24. Zhang Q, Miao Y, Fu Q, et al. CircRNACCDC66 regulates cisplatin resistance in gastric cancer via the miR-618/BCL2 axis. Biochem Biophys Res Commun. 2020;526(3):713–720.
25. Yang W, Ju HY, Tian XF. Circular RNA-ABCB10 suppresses hepatocellular carcinoma progression through upregulating NRP1/ABL2 via sponging miR-340-5p/miR-452-5p. Eur Rev Med Pharmacol Sci. 2020;24(5):2347–2357.
26. He Z, Xia Y, Pan C, et al. Up-regulation of MiR-452 inhibits metastasis of non-small cell lung cancer by regulating BMI1. Cell Physiol Biochem. 2015;37(1):387–398. doi:10.1159/000430362
27. Zhang Y, Han L, Pang J, Wang Y, Feng F, Jiang Q. Expression of microRNA-452 via adenoviral vector inhibits non-small cell lung cancer cells proliferation and metastasis. Tumour Biol. 2016;37(6):8259–8270. doi:10.1007/s13277-015-4725-z
28. Song N, Liu B, Wu JL, et al. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34(5):2599–2603. doi:10.1007/s13277-013-0807-y
29. Song X, Wang H, Wu J, Sun Y. Long noncoding RNA SOX2-OT knockdown inhibits proliferation and metastasis of prostate cancer cells through modulating miR-452-5p/HMGB3 axis and inactivating wnt/beta-catenin pathway. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2019.3479
30. Shi J, Wang H, Feng W, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol. 2019;862:172615. doi:10.1016/j.ejphar.2019.172615
31. Xie Y, Wang L, Yang D. CircEPSTI1 promotes the progression of non-small cell lung cancer through miR-145/HMGB3 axis. Cancer Manag Res. 2020;12:6827–6836. doi:10.2147/CMAR.S252893
32. Zhou ZF, Wei Z, Yao JC, et al. CircRNA_102179 promotes the proliferation, migration and invasion in non-small cell lung cancer cells by regulating miR-330-5p/HMGB3 axis. Pathol Res Pract. 2020;216(11):153144. doi:10.1016/j.prp.2020.153144
33. Liu C, Li H, Jia J, Ruan X, Liu Y, Zhang X. High metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) expression promotes proliferation, migration, and invasion of non-small cell lung cancer via ERK/Mitogen-activated protein kinase (MAPK) signaling pathway. Med Sci Monit. 2019;25:5143–5149. doi:10.12659/MSM.913308
34. He C, Bai X, Li Y, et al. Runt-related transcription factor 1 contributes to lung cancer development by binding to tartrate-resistant acid phosphatase 5. Cell Cycle. 2019;18(23):3404–3419. doi:10.1080/15384101.2019.1678966
35. Cao Q, Mao ZD, Shi YJ, et al. MicroRNA-7 inhibits cell proliferation, migration and invasion in human non-small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget. 2016;7(47):77468–77481. doi:10.18632/oncotarget.12684
36. Liu J, Wang L, Li X. HMGB3 promotes the proliferation and metastasis of glioblastoma and is negatively regulated by miR-200b-3p and miR-200c-3p. Cell Biochem Funct. 2018;36(7):357–365. doi:10.1002/cbf.3355
37. Choi HW, Manohar M, Manosalva P, Tian M, Moreau M, Klessig DF. Activation of plant innate immunity by extracellular high mobility group box 3 and its inhibition by salicylic acid. PLoS Pathog. 2016;12(3):e1005518. doi:10.1371/journal.ppat.1005518
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.