Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA 0001073 Attenuates Malignant Biological Behaviours in Breast Cancer Cell and Is Delivered by Nanoparticles to Inhibit Mice Tumour Growth
Authors Yi Z, Li Y, Wu Y , Zeng B, Li H, Ren G , Wang X
Received 7 February 2020
Accepted for publication 16 June 2020
Published 29 June 2020 Volume 2020:13 Pages 6157—6169
DOI https://doi.org/10.2147/OTT.S248822
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Ziying Yi,1,2 Yunhai Li,1,2 Yushen Wu,1 Beilei Zeng,1 Hongzhong Li,1,2 Guosheng Ren,1,2 Xiaoyi Wang2
1Chongqing Key Laboratory of Molecular Oncology and Epigenetics, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Department of Endocrine and Breast Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Guosheng Ren; Xiaoyi Wang
The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, People’s Republic of China
Tel +86 023 8901 1477
Fax +86 023 8901 2305
Email [email protected]; [email protected]
Background: Circular RNAs (circRNAs) are a special class of noncoding RNAs that are involved in gene regulation and compete with mRNA for miRNA binding sites. The roles of circRNAs in cancer, especially breast cancer (BC), are poorly understood.
Materials and Methods: The expression levels of circRNA 0001073 (circ-1073) in BC cells (BCCs) and tissues and peritumoural tissues were detected by real-time quantitative reverse transcription-polymerase chain reaction. Kaplan–Meier analysis and receiver operating characteristic curves were used to evaluate relapse-free survival (RFS) and the diagnostic value of circ-1073 for BC, respectively. The biological functions of circ-1073 were determined by cell counting kit-8 assays, colony formation assays, flow cytometry, wound-healing assays, transwell assays, and xenograft model studies. RNA immunoprecipitation assays were conducted to identify the connection between circ-1073 and human antigen R (HuR).
Results: Low circ-1073 expression was discovered in BCCs and BC tissues compared with normal mammary epithelial cells and peritumoural tissues, respectively. Circ-1073 downregulation was significantly associated with an unfavourable prognosis, including a shorter RFS, in BC patients. Circ-1073 is a valuable diagnostic biomarker for BC. Circ-1073 overexpression significantly inhibited BCC proliferation and induced apoptosis by increasing Cleaved Caspase-3/9 levels. Moreover, circ-1073 upregulation significantly suppressed cell mobility and epithelial–mesenchymal transition. Notably, xenograft tumour growth was inhibited by the intratumoural injection of nanoparticles containing the circ-1073 plasmid or by circ-1073 overexpression, and this inhibition was accompanied by HuR upregulation.
Conclusion: Circ-1073 functions as a tumour suppressor in BC, suggesting its potential as a novel therapeutic target in BC.
Keywords: circ-1073, breast cancer, apoptosis, metastasis, nanoparticles, HuR
Introduction
Globally, breast cancer (BC) ranks first in female cancer-related morbidity and is the second-leading threat to the lives of women.1 Fortunately, numerous markers and therapeutic strategies have been discovered for more precise and effective BC treatment. Nonetheless, more work is needed.
Circular RNAs (circRNAs) belong to a special class of noncoding RNAs with regulatory potential2,3 and are broadly expressed across cells and tissues.4 CircRNAs form single-stranded circular structures by covalent binding;5,6 these molecules are more stable than linear RNAs and show greater resistance to the exonuclease RNase R.7,8 Multiple circRNAs have been shown to be more abundant (> 10-fold higher) than their linear RNAs.9,10 However, circRNAs are usually expressed at low levels,11,12 suggesting they probably play a regulatory role in splicing noise.5 CircRNAs in nuclei modulate the transcription of their parental genes in combination with RNA polymerase II.13 Additionally, many elegant studies have proven that circRNAs have modulatory function in multiple cancers, including gastric carcinoma,14,15 urothelial carcinoma,16,17 bronchogenic carcinoma,18 and renal cell carcinoma,19 with some circRNAs serving as tumour promoters and others as tumour suppressors. Although some knowledge of the characteristics and mechanism of circRNA formation is available, the roles of circRNAs in malignant tumours, especially BC, remain unclear.
Activin A receptor type 2A (ACVR2A), a member of the transforming growth factor-beta superfamily, plays an important role in mediating the function of activins.20 ACVR2A expression was previously found to be downregulated in colon carcinoma, which was related to an unfavourable prognosis.21 However, the roles of circular ACVR2A in BC remain unknown.
In our study, we first identified circRNA 0001073 (circ-1073) derived from the ACVR2A gene as a regulator in BC. We detected circ-1073 expression in BC tissues and BC cells (BCCs) and then analysed the clinical features of BC patients. Importantly, we demonstrated the biological functions of circ-1073 in BCCs. A brilliant previous study developed a nanoparticle-based siRNA transfer system that can be applied to intratumoural injections for cancer therapy.22 We then used a nano-transfection reagent to control tumour growth in vivo. Interestingly, we discovered an interaction between circ-1073 and human antigen R (HuR; a typical RNA-binding protein, RBP), which is known to inhibit cancer cell growth and motility by binding circRNAs.23,24 HuR overexpression suppresses BC progression by decreasing vascular endothelial growth factor alpha (VEGFα) stability.25 Therefore, we hypothesized that circ-1073 binds HuR to regulate the expression levels of downstream targets to influence the malignant biological behaviour of tumour cells. In conclusion, circ-1073 may prove a novel strategy for the therapy of BC.
Materials and Methods
Mammary Tissues
In total, 112 paired BC tissues and peritumoural tissues were obtained from surgical specimens from patients with BC at the First Affiliated Hospital of Chongqing Medical University. The clinical specimens were confirmed by pathological examination. The experiment was approved by the Ethics Committees of the First Affiliated Hospital of Chongqing Medical University. All patients provided written informed consent which complied with the 1964 the Declaration of Helsinki.
Cell Lines
Human BCCs (MDA-MB-231, MDA-MB-468, BT-549, MCF-7, T47D, ZR-75-1, and SK-BR-3) and human normal breast epithelial MCF-10A cells were obtained from American Type Culture Collection (ATCC). BCCs were maintained in RPMI 1640 medium with 10% foetal bovine serum (FBS; Gibco, Carlsbad, CA). MCF-10A cells were maintained as described previously.26 All cells were cultured at 37 °C in a 5% CO2 incubator.
Transfection and Stable Cell Line Construction
The human circ-1073 linear sequence (473 bp) was synthesized and cloned into pcDNA3.1 (a mammalian expression plasmid) by GeneCopoeia (Rockville, MD, USA) to construct overexpression plasmids. Plasmids were delivered into cells with Lipofectamine™ 3000 transfection reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s protocols. Stable cell lines were obtained by geneticin (G418) selection for 14 days.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from tissues and cell samples with TRIzol Reagent (Invitrogen, Carlsbad, CA). RNA concentration was detected using a Thermo Nanodrop 2000 spectrophotometer (Waltham, MA). RNA quality was evaluated according to the A260/A280 ratio. RNA was reverse transcribed to produce cDNA with a Reverse Transcription Kit (Promega, Madison, WI). RT-qPCR was performed using GoTaq SYBR Green (Promega, Madison, WI) with a CFX96 Touch Deep Well RT-PCR System (Bio-Rad, Hercules, CA). The specific primers are as follows: β-actin (forward: 5ʹ-CTCTGCCCGCATGAACCT-3ʹ, reverse: 5ʹ-CCACCATCCACATCCCAC-3ʹ) and circ-1073 (forward: 5ʹ-AGTCAGTTCCTTGTGGAGCC-3ʹ, reverse: 5ʹ-GCATGGGTTCTGACGGACAT-3ʹ). β-Actin was amplified as the reference standard for normalization.
Fluorescence in situ Hybridization (FISH)
FISH was conducted as previously described.27 A FITC-labelled circ-1073 probe was constructed by Bersinbio (Guangzhou, China) with the sequence 5ʹ-TACCAAGTATAGCACTTGAGTTGGAACAAG-3ʹ. Circ-1073 signals were detected with a FISH Kit (Bersinbio, Guangzhou, China) following the manufacturer’s detailed instructions; nuclei were stained with 4ʹ,6-diamidino-2-phenylindole (DAPI). All photos were captured with an LSM 800 confocal microscope system (Zeiss, Germany).
Cell Proliferation
Cell proliferation was evaluated at specific time points with Cell Counting Kit-8 (CCK8; Beyotime, Shanghai, China). BCCs were counted and plated into 96-well plates, and the absorbance was detected at 450 nm using a microplate reader (Infinite 200 PRO, TECAN, Männedorf, Switzerland). Stable transfectants were plated at 1000 cells/well into 6-well plates in triplicate for colony formation assays. After ten days, colonies were rinsed gently three times with phosphate-buffered saline (PBS), fixed in methanol and stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 15 min. The colonies were then imaged and quantified.
Apoptosis Analysis
Apoptosis was assessed using the Annexin V-FITC Apoptosis Detection Kit (Invitrogen, Carlsbad, CA). First, the cells were rinsed once in PBS and once in 1X Binding Buffer. The cells were then suspended in a mixture containing 5 μL of FITC-Annexin V and 100 μL of 1X Binding Buffer and incubated for 15 min at room temperature. After being rinsed in 1X Binding Buffer, the cells were resuspended in a mixture of 5 μL of propidine iodide and 200 μL of 1X Binding Buffer. The data collected with a FACSCanto flow cytometer (BD, Franklin Lakes, NJ) were analysed by FlowJo 10.
Cell Migration and Invasion
Cell motility was assessed by wound-healing assays as previously described.28 After a wound was created with a sterile 10-μL tip, the cells were maintained in serum-free medium and photographed by phase-contrast microscopy (Leica DMI4000B, Bucks, UK) at specific time points. An 8-μm pore size transwell chamber (Corning, Bedford, MA) was utilized for cell migration assays, and Matrigel (BD, Franklin Lakes, NJ) was added to these chambers for cell invasion assays. Approximately 2 × 104 cells resuspended in 100 μL of serum-free RPMI 1640 medium were plated in the upper chamber, and approximately 600 μL of medium with 10% FBS was placed in the lower chamber as an artificial chemoattractant. After staining, the cells on the underside of the membrane were imaged, and the migration and invasion results were statistically analysed.
Western Blotting (WB)
Proteins were isolated from samples using RIPA lysis buffer (Thermo Scientific, Waltham, MA) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Protein concentration was determined and WB was conducted as previously described.28 The primary antibodies were as follows: anti-β-Tubulin (#86,298), anti-Cleaved Caspase-9 (#9501), anti-Caspase 9 (#9502), anti-Cleaved Caspase-3 (#9661), anti-Caspase 3 (#9662), anti-E-cadherin (#3195), anti-Vimentin (#574), and anti-MMP9 (#3852) (Cell Signaling Technology, Danvers, MA).
Immunofluorescence (IF)
Cells cultured overnight on slides were rinsed three times with PBS and fixed in methanol for 15 min at room temperature. After permeabilization with PBS containing 0.1% Triton X-100, the cells were rinsed with PBS and blocked in 5% goat serum for 20 min at room temperature. The cells were then incubated with anti-E-cadherin or anti-Vimentin antibody overnight at 4 °C, rinsed with PBS, and incubated with fluorescence-labelled secondary antibodies for 0.5–1 h at 37 °C. The nuclei were counterstained with DAPI for 5 min at room temperature. The cells were rinsed with PBS and photographed using an LSM 800 confocal microscope system (Zeiss, Germany).
Xenograft Tumours and Micropoly (MP)-Circ-1073 Treatment
Xenograft tumour models were established in 6-week-old female nude mice upon the subcutaneous injection of suspensions containing 5 × 106 MDA-MB-231-circ-1073 or control cells (n = 5 per group). Tumours were measured, and tumour volume was calculated using the classic formula ½ (length × width2). Micropoly-transfecterTM Tissue Reagent (MT215, Micropoly Biotech, Shenzhen, China), a highly efficient nano-transfection reagent, was utilized for circ-1073 treatment. The mice received an intratumoural injection of MP-control or MP-circ-1073 every 5 days for 1 month following the manufacturer’s instructions. The mixture injected into each mouse consisted of 15 μL of MP and 15 μg of plasmid. The experiment has obtained approval by the Institutional Ethics Committees of the First Affiliated Hospital of Chongqing Medical University and were carried out following the guidelines and regulations of the Animal Care and Use Committees of the First Affiliated Hospital of Chongqing Medical University. All mice were administered standard feed and kept in specific pathogen-free house on a 12 h light–dark cycle. Temperature was maintained at 24°C±2°C and humidity was kept at 50%±5%.
Immunohistochemistry (IHC)
IHC was performed as previously described.29 After incubation with anti-Cleaved Caspase-3 or anti-HuR antibodies (#12,582) overnight at 4 °C, the samples were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 10 min at 37 °C. All imaged were obtained using an upright microscope (BX53, Olympus, Japan).
RNA Immunoprecipitation (RIP)
RIP assays were performed according to the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA) protocols. Coprecipitated RNAs were quantified by RT-PCR with a specific primer for ELAVL1 (HuR coding gene). Total RNA (input) was used as a control.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 5.0 (San Diego, CA). Low circ-1073 expression was defined as cases with a Ct value greater than the median (8.091); high circ-1073 expression was defined as cases with a Ct value less than the median. Data are presented as the mean ± standard error of the mean (SEM) or the mean ± standard deviation (SD) of three independent experiments. Kaplan–Meier survival curves were compared by the Log rank test. Circ-1073 levels in BC were analysed to generate receiver operating characteristic (ROC) curves; the diagnostic value of circ-1073 was evaluated by the area under the curve (AUC). Results with a p-value < 0.05 were classified as statistically significant. Differences between groups were analysed with two-tailed Student’s t-tests.
Results
Circ-1073 Expression Is Downregulated in BCCs
As recorded in the cancer-specific circRNA database (CSCD)4 (http://gb.whu.edu.cn/CSCD/), 11 circRNAs originate from the human ACVR2A gene, 6 of which are expressed in cancer cells; however, only circ-1073 was expressed in MCF-7 cells and downregulated in BCCs compared with MCF-10A cells (Figure 1A). According to the RT-qPCR analysis, circ-1073 expression was clearly lower in BCCs than in MCF-10A cells (p < 0.001, Figure 1B). Circ-1073 is located on chromosome 2, and Sanger sequencing of the 473-nt circ-1073 was conducted to confirm head-to-tail splicing before cyclization (Figure 1C). As covalent circularization renders an RNA highly resistant to exonuclease digestion, circ-1073 was more resistant to RNase R treatment than linear ACVR2A or β-actin, as determined by RT-qPCR (p < 0.001, Figure 1D). Overall, these findings indicate that circ-1073 is derived from the ACVR2A gene and that circ-1073 expression is downregulated in BCCs.
 |
Figure 1 Circ-1073 expression is downregulated in breast cancer cells (BCCs). (A) Circ-1073 is predicted to be broadly expressed in diverse tumour cells, including MCF-7 cells, by the CSCD (http://gb.whu.edu.cn/CSCD/). (B) Circ-1073 expression was downregulated in 7 BCCs compared with MCF10A cells, as determined by RT-qPCR (p < 0.001). (C) A schematic illustration shows the genomic location of circ-1073 in its host gene, which was validated by Sanger sequencing. (D) Circ-1073 was at least 4-times more resistant to RNase R than linear ACVR2A, as determined by RT-qPCR (p < 0.001). Mean ± SD. ns = not significant; ***p < 0.001 by Student’s t-test. Abbreviations: BCCs, breast cancer cells; CSCD, cancer-specific circRNA database; RT-qPCR, Real-time quantitative polymerase chain reaction. |
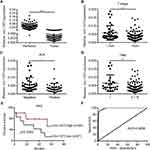
Circ-1073 Expression Is Downregulated in BC Tissues
Circ-1073 expression was lower in BC tissues than in peritumoural mammary tissues (p < 0.001) (Figure 2A). The clinical relevance of circ-1073 expression levels was analysed in 132 BC patients, and the results revealed that circ-1073 was strongly related to age, tumour size, axillary lymph node (ALN) status, and preoperative chemotherapy status (Table 1). Low circ-1073 expression was significantly associated with large tumours (p = 0.006, Figure 2B), ALN positivity (p = 0.014, Figure 2C), and advanced Tumor Node Metastasis (TNM) stage (p = 0.009, Figure 2D). Furthermore, Kaplan–Meier survival curves indicated an unfavourable relapse-free survival (RFS) in BC patients with low circ-1073 expression (p = 0.038, Figure 2E). A ROC curve was constructed to assess the potential of circ-1073 as a biomarker for BC patients, and the AUC was calculated to be 0.9898 (95% CI 0.9818 to 0.9979, p < 0.001) (Figure 2F); the optimal cut-off value was 6.68 with 92.42% sensitivity and 97.37% specificity. These data show that circ-1073 expression is downregulated in BC tissues and that circ-1073 may be a new diagnostic biomarker for BC.
 |
Table 1 Analysis of the Relationship Between Circ-1073 Level and Clinicopathological Factors |
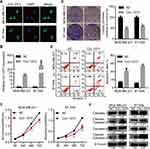
Circ-1073 Suppresses BCCs Proliferation and Promotes Apoptosis
To explore more characteristics of circ-1073, FISH was conducted to determine the cytoplasmic and nuclear localization of circ-1073 in MDA-MB-231 and BT-549 cells (Figure 3A). To reveal the biological roles of circ-1073 in BCCs, MDA-MB-231 and BT-549 cells, which have the lowest circ-1073 expression among BCCs, were transfected with the pcDNA3.1-circ-1073 vector to establish overexpression cell lines (indicated by circ-1073); the pcDNA3.1 plasmid was used as a negative control (NC) (p < 0.01, Figure 3B). CCK8 assays showed the reduced viability of circ-1073-overexpressing cells compared with NC cells (p < 0.001, Figure 3C). In addition, colony formation was markedly inhibited by circ-1073 overexpression (p < 0.001, Figure 3D). Furthermore, flow cytometry showed increased apoptosis of circ-1073-overexpressing cells (p < 0.05, Figure 3E). Many elegant studies have shown that increased Cleaved Caspase-3/9 levels promote cell apoptosis.30–32 Based on WB, the levels of total Caspase 3/9 were unchanged, but Cleaved Caspase-3/9 accumulated in the circ-1073 group compared with the NC group (p < 0.001, Figure 3F). In general, the data show that circ-1073 inhibits proliferation and induces apoptosis, accompanied by elevated levels of Cleaved Caspase-3/9 in circ-1073-overexpressing cells compared with control cells.
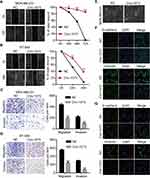
Circ-1073 Reduces BCCs Migration and Invasion and Reverses Epithelial–Mesenchymal Transition (EMT)
To further elucidate the biological function of circ-1073, wound-healing assays were conducted, which demonstrated the weakened motility of circ-1073-overexpressing MDA-MB-231 (p = 0.003, Figure 4A) and BT-549 cells (p = 0.003, Figure 4B). As shown by Transwell assays, circ-1073 overexpression significantly reduced the migration and invasion of MDA-MB-231 (p < 0.001, Figure 4C) and BT-549 cells (p < 0.001, Figure 4D). Scientists usually characterize MDA-MB-231 cells as highly migratory and invasive mesenchymal-like cells33 with low E-cadherin expression but high Vimentin expression.34 Notably, phase contrast microscopy showed a morphological change in MDA-MB-231 cells from a mesenchymal-like phenotype to an epithelial-like phenotype upon circ-1073 overexpression (Figure 4E), implying weakened EMT. Therefore, we detected EMT marker levels by IF and discovered increased E-cadherin expression but decreased Vimentin expression in circ-1073-overexpressing MDA-MB-231 (Figure 4F) and BT-549 cells (Figure 4G). Collectively, these findings show that circ-1073 overexpression hinders BCC migration and invasion, upregulates E-cadherin expression, and downregulates Vimentin expression.
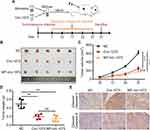
Circ-1073 Effectively Delays Xenograft Tumour Growth
Stable MDA-MB-231 cell lines overexpressing circ-1073 or NC were subcutaneously injected into female nude mice, and mice injected with NC cells were randomized into the control and MP-circ-1073 groups. As an innovative approach, the mice in the MP-circ-1073 group received intratumoural injections of circ-1073 in a mixture with MP (Figure 5A); the control mice were injected with MP alone. Interestingly, tumour growth was significantly impeded in the circ-1073-overexpressing and MP-circ-1073 groups compared with the control groups (p < 0.05, Figure 5B–D). Since WB showed increased Cleaved Caspase-3 levels in BCCs, IHC was performed, and Cleaved Caspase-3 levels were clearly elevated in the circ-1073-overexpressing and MP-circ-1073 groups (p < 0.05, Figure 5E). Studies have revealed that circ-1073 suppresses tumorigenesis.
The RBP HuR Interacts with Circ-1073
To explore the mechanism by which circ-1073 impedes tumour progression, two databases, CircInteractome (https://circinteractome.nia.nih.gov/) and CSCD, were employed to predict RBPs and binding sites. Surprisingly, HuR was predicted to be the RBP for circ-1073. The results of analysis using catRAPID, another database (http://service.tartaglialab.com/page/catrapid_group), showed that circ-1073 potentially interacts with HuR (Figure 6A), and RIP was performed to confirm binding between circ-1073 and HuR (Figure 6B). Notably, HuR expression was markedly higher in the circ-1073 and MP groups than in the NC group (p < 0.01, Figure 6C). Our findings illustrate that HuR levels decrease through binding overexpressed circ-1073. The conclusions of our in vitro and in vivo experiments are summarized in Figure 7.
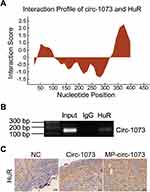 |
Figure 6 Circ-1073 binds to HuR. (A) HuR interacts with circ-1073, as shown in the catRAPID database (http://www.tartaglialab.com/). (B) Binding between circ-1073 and HuR was verified by RIP assays. (C) HuR expression was significantly upregulated in the circ-1073 and MP groups compared with the control group (p < 0.01). Abbreviations: HuR, human antigen R; IgG, immunoglobulin G; RIP, RNA immuno precipitation; NC, negative control; MP, micropoly. |
Discussion
CircRNAs were discovered in the cytoplasm of eukaryotic cells and defined as transcripts resulting from splicing errors.35,36 Later, circRNAs were found in large numbers by high-throughput sequencing.37 Emerging studies have revealed the features of circRNAs in mammalian cells and have shown that they are derived from exons,38 introns,39 or exon-intron regions.40 Most circRNAs, including circ-1073, originate from exons and are covalently closed RNA molecules produced by back-splicing.41 Despite the widespread expression of circRNAs in most tissues, an influential study accurately measured circRNA levels in normal and cancerous tissues and found that circRNAs were commonly expressed at lower levels in highly dividing cells, particularly tumour cells.42 After predicting the distribution of circ-1073 in MCF-7 cells, we discovered reduced circ-1073 expression in BC and hypothesized that this decrease may be responsible for the malignant behaviour of BCCs. In our study, low circ-1073 expression was linked to poor clinical features and thus may be a diagnostic marker for BC. As expected, our work indicates that circ-1073 significantly slows the growth of BCCs. Previous research has indicated that circRNAs not only suppress cancer cell proliferation but also inhibit migration and invasion;43,44 we had similar observations, ie, circ-1073 attenuated the aggressive behaviours of BCCs. Surprisingly, we found that circ-1073 reverted BCCs from a mesenchymal phenotype to an epithelial phenotype.
After observing the antitumour effects in vitro, we next explored the function of circ-1073 in vivo and detected a marked decrease in tumour growth rate. In many studies, the target gene is edited in cells that are then injected to generate subcutaneous tumours; these modified cells often show delayed tumour growth. Nonetheless, we innovatively applied Micropoly-transfecter, a plasmid delivery system, to deliver the circ-1073 plasmid by intratumoural injection to inhibit tumour progression. Nanoparticles have been applied in many ways, such as intravenous injection45 and tail vein injection,46 to treat tumours, and nanoparticles are a promising tool for cancer treatment. To initially determine the mechanism of tumour suppression by circ-1073, CircInteractome was queried, and HuR was predicted as the RBP that interacts with circ-1073. In our work, elevated HuR expression was achieved by circ-1073 overexpression in vivo. As reported in various articles, circRNA cyclization is mediated by the binding of RBPs to introns flanking the circRNA-encoding exons.47 Therefore, HuR likely binds circ-1073 to promote its cyclization and exert biological functions. Regrettably, we have not thoroughly explored the molecular features and mechanism of the connection between circ-1073 and HuR.
A previous study reported that circRNA-100,269 levels were downregulated in gastric carcinoma and negatively related to those of miR-630; circRNA-100,269 overexpression evidently suppresses tumour cell proliferation.48 Another study showed that circRNA-LARP4 impedes gastric cancer cell growth and motility as a miR-424-5p sponge.49 Notably, a recent study published in Molecular Cancer identified circ-1073 as a promising tumour suppressor in bladder cancer, stating that circ-1073 suppresses bladder cancer cell proliferation and metastasis by sponging miR-626.50 All of these previous studies mentioned the classic mechanism of circRNA function, namely, circRNAs act as miRNA sponges through mechanical binding. With the development of bioinformatics, this approach is increasingly used for data mining in studies on circRNAs. As shown by CircInteractome, 15 miRNAs potentially interact with circ-1073. However, we did not delve deeply into the relationships between circ-1073 and its miRNAs.
Conclusion
Our research elucidated not only the expression profile and clinical importance of circ-1073 in BC but also the biological effects and preliminary mechanism through which circ-1073 impedes BC progression. In an innovative approach, we used MP to introduce circ-1073 for the treatment of BC in a mouse model. We deduce that circ-1073 binds to HuR and increases HuR expression to inhibit the malignant biological behaviour of BCCs by increasing cleaved-Caspase 3/9 and E-cadherin levels. However, further work is necessary to clarify the detailed molecular mechanism of circ-1073. Nano-based MP delivery of circ-1073 is a potential candidate strategy for BC treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
3. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
4. Xia S, Feng J, Chen K, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi:10.1093/nar/gkx863
5. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-6
6. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
7. Dang Y, Yan L, Hu B, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17(1):130. doi:10.1186/s13059-016-0991-3
8. Ruan H, Xiang Y, Ko J, et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 2019;11(1):55. doi:10.1186/s13073-019-0663-5
9. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO, Moran JV. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi:10.1371/journal.pgen.1003777
10. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. doi:10.1038/ncomms11215
11. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
12. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi:10.1186/s13059-014-0409-z
13. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi:10.1016/j.canlet.2018.03.035
14. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi:10.1038/bjc.2016.451
15. Chen Y, Yang F, Fang E, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26(7):1346–1364. doi:10.1038/s41418-018-0220-6
16. Zeng Z, Zhou W, Duan L, et al. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol. 2019;234(4):3887–3896. doi:10.1002/jcp.27162
17. Chen X, Chen RX, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–6330. doi:10.1158/1078-0432.Ccr-18-1270
18. Tan S, Sun D, Pu W, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. 2018;17(1):138. doi:10.1186/s12943-018-0887-9
19. Franz A, Ralla B, Weickmann S, et al. Circular RNAs in clear cell renal cell carcinoma: their microarray-based identification, analytical validation, and potential use in a clinico-genomic model to improve prognostic accuracy. Cancers (Basel). 2019;11(10):1473. doi:10.3390/cancers11101473
20. Wang SS, Huang HY, Chen SZ, et al. Gdf6 induces commitment of pluripotent mesenchymal C3H10T1/2 cells to the adipocyte lineage. FEBS J. 2013;280(11):2644–2651. doi:10.1111/febs.12256
21. Zhuo C, Hu D, Li J, et al. Downregulation of activin A receptor type 2A is associated with metastatic potential and poor prognosis of colon cancer. J Cancer. 2018;9(19):3626–3633. doi:10.7150/jca.26790
22. Li X, Chen Y, Wang M, et al. A mesoporous silica nanoparticle–PEI–fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials. 2013;34(4):1391–1401. doi:10.1016/j.biomaterials.2012.10.072
23. Latorre E, Carelli S, Raimondi I, et al. The ribonucleic complex HuR-MALAT1 represses CD133 expression and suppresses epithelial-mesenchymal transition in breast cancer. Cancer Res. 2016;76(9):2626–2636. doi:10.1158/0008-5472.CAN-15-2018
24. Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi:10.1080/15476286.2017.1279788
25. Gubin MM, Calaluce R, Davis JW, et al. Overexpression of the RNA binding protein HuR impairs tumor growth in triple negative breast cancer associated with deficient angiogenesis. Cell Cycle. 2010;9(16):3337–3346. doi:10.4161/cc.9.16.12711
26. Halsne R, Tandberg JI, Lobert VH, et al. Effects of perfluorinated alkyl acids on cellular responses of MCF-10A mammary epithelial cells in monolayers and on acini formation in vitro. Toxicol Lett. 2016;259:95–107. doi:10.1016/j.toxlet.2016.08.004
27. El Gammal AT, Bruchmann M, Zustin J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16(1):56–64. doi:10.1158/1078-0432.CCR-09-1423
28. Feng Y, Feng L, Yu D, Zou J, Huang Z. srGAP1 mediates the migration inhibition effect of Slit2-Robo1 in colorectal cancer. J Exp Clin Cancer Res. 2016;35(1):191. doi:10.1186/s13046-016-0469-x
29. Aguado A, Rodriguez C, Martinez-Revelles S, et al. HuR mediates the synergistic effects of angiotensin II and IL-1β on vascular COX-2 expression and cell migration. Br J Pharmacol. 2015;172(12):3028–3042. doi:10.1111/bph.13103
30. Quintana C, Cabrera J, Perdomo J, et al. Melatonin enhances hyperthermia-induced apoptotic cell death in human leukemia cells. J Pineal Res. 2016;61(3):381–395. doi:10.1111/jpi.12356
31. Chen S, Li H, Zhuang S, et al. Sam68 reduces cisplatin-induced apoptosis in tongue carcinoma. J Exp Clin Cancer Res. 2016;35(1):123. doi:10.1186/s13046-016-0390-3
32. Zhang X, Li S, Dong C, Xie X, Zhang Y. Knockdown of long noncoding RNA NR_026689 inhibits proliferation and invasion and increases apoptosis in ovarian carcinoma HO-8910PM cells. Oncol Res. 2017;25(2):259–265. doi:10.3727/096504016X14732503870766
33. Sero JE, Sailem HZ, Ardy RC, et al. Cell shape and the microenvironment regulate nuclear translocation of NF-κB in breast epithelial and tumor cells. Mol Syst Biol. 2015;11(3):790. doi:10.15252/msb.20145644
34. Galarza TE, Mohamad NA, Taquez Delgado MA, et al. Histamine prevents radiation-induced mesenchymal changes in breast cancer cells. Pharmacol Res. 2016;111:731–739. doi:10.1016/j.phrs.2016.07.039
35. Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi:10.1016/0092-8674(93)90279-Y
36. Schindewolf CA, Domdey H. Splicing of a circular yeast pre-mRNA in vitro. Nucleic Acids Res. 1995;23(7):1133–1139. doi:10.1093/nar/23.7.1133
37. Hoffmann S, Otto C, Doose G, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15(2):R34. doi:10.1186/gb-2014-15-2-r34
38. Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218. doi:10.1186/s13059-018-1594-y
39. Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29(20):2168–2182. doi:10.1101/gad.270421.115
40. Li Z, Huang C, Bao C, et al. Corrigendum: exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2017;24(2):194. doi:10.1038/nsmb0217-194a
41. Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–1287. doi:10.1101/gr.202895.115
42. Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5(1):8057. doi:10.1038/srep08057
43. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. doi:10.1002/1878-0261.12045
44. Lin Q, Ling YB, Chen JW, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol. 2018;53(1):246–256. doi:10.3892/ijo.2018.4371
45. Christie RJ, Matsumoto Y, Miyata K, et al. Targeted polymeric micelles for siRNA treatment of experimental cancer by intravenous injection. ACS Nano. 2012;6(6):5174–5189. doi:10.1021/nn300942b
46. Haque F, Shu D, Shu Y, et al. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7(4):245–257. doi:10.1016/j.nantod.2012.06.010
47. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi:10.1038/nrm.2015.32
48. Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging. 2017;9(6):1585–1594. doi:10.18632/aging.101254
49. Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi:10.1186/s12943-017-0719-3
50. Dong W, Bi J, Liu H, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18(1):95. doi:10.1186/s12943-019-1025-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.