Back to Journals » Cancer Management and Research » Volume 13
circTADA2A Retards the Progression of Colorectal Cancer via Regulating miR-1229/BCL2L10 Signal Axis
Authors Hu L, Fang L, Zhang Z, Yan Z
Received 6 April 2021
Accepted for publication 6 August 2021
Published 1 September 2021 Volume 2021:13 Pages 6811—6821
DOI https://doi.org/10.2147/CMAR.S314548
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
Li Hu, Lei Fang, Zhiping Zhang, Zhilong Yan
Department of Gastroenterology, Ningbo No.1 Hospital, Ningbo, People’s Republic of China
Correspondence: Zhilong Yan
Department of Gastroenterology, Ningbo No.1 Hospital, 59 Liuting Street, Haishu District, Ningbo, 315000, Zhejiang Province, People’s Republic of China
Email [email protected]
Background: Colorectal cancer (CRC) is one of the leading causes of cancer-related death around the world, becoming a severe public health problem. Mounting evidence has proven that circRNAs act as pivotal modulators in the initiation and development of CRC. Although the function of circTADA2A has been explored in osteosarcoma and breast cancer, the specific role of circTADA2A in CRC remains unknown.
Methods: Bioinformatics analysis based on GEO datasets was used to evaluate the dysregulated circRNAs in CRC. CCK-8 and transwell assays were used to detect the functions of CRC cells. qRT-PCR and Western blot were performed to evaluate the expression of RNAs and proteins. Luciferase assay and RNA pull down experiment were carried out to verify the interaction between miR and its targets.
Results: CircTADA2A was downregulated in CRC tissues compared with normal samples. CircTADA2A exhibited greater stability than its linear form when exposed to RNase R and actinomycin D treatment. qRT-PCR analysis validated the lower expression level of circTADA2A in CRC. The loss-of-function and gain-of-function assays indicated that circTADA2A exerted the inhibitory role in CRC cell proliferation and migration. Mechanistically, circTADA2A functioned as a sponge of miR-1229. Further experiments manifested that circTADA2A regulated BCL2L10 expression via competitively binding to miR-1229. More importantly, the tumor suppressor role of circTADA2A in the malignant behaviors of CRC cells was mediated by BCL2L10.
Conclusion: circTADA2A suppressed cell proliferation and migration in CRC through regulation of miR-1229/BCL2L10 axis, which suggested that circTADA2A might represent a novel potential target for the treatment of CRC.
Keywords: colorectal cancer, circTADA2A, miR-1229, BCL2L10
Introduction
Reportedly, colorectal cancer (CRC) ranks third among the prevalent malignant tumors and is one of the leading contributors responsible for cancer-related mortality globally.1,2 In recent decades, the incidence of CRC is sharply increasing attributable to changes in diet and lifestyle, triggering a serious threat to human health.3 The lethality of CRC stabilizes at a high level for the reason that the characteristics of early CRC are unconspicuous and patients are generally diagnosed at an advanced stage of this disease.4 Nevertheless, if patients are in the early stages of CRC when diagnosed, it is estimated that 90% of deaths may be avertible.5 Accordingly, gaining a deep understanding of the pathogenic mechanism governing CRC is urgently needed to improve early diagnosis and clinical intervention for CRC.
Recently, noncoding RNAs have attracted the extensive attention of scientists due to their important roles in the modulation of diverse cellular processes.6 Growing evidence indicates that dysfunction of ncRNAs is considered as a key factor in the pathological development of various disorders.7,8 As a novel type of endogenous conserved noncoding RNA transcripts, circular RNAs (circRNAs) are featured with a circular configuration generated by covalent binding of 3’and 5’ ends via back-splicing.9,10 Therefore, circRNAs are evolutionarily conserved and highly stable as opposed to linear RNAs with 5’ caps and 3’ tails owing to the deficiency of 5′ to 3′ polarity or poly-A tail in closed-loop structures.11 Abundant circRNAs are reported to display specific expression in tissues as well as developmental stages and act as regulators in the tumorigenesis and progression of multiple cancers, including CRC.12–14 circRNAs are participated in the pathogenesis of CRC through regulating the expression of core target genes via different mechanisms, such as sponging microRNA (miRNA), working as protein scaffolds, modulating alternative splicing, transcription and RNA-protein interplays.15–18 In view of this, it is essential to identify the characteristics of circRNAs in CRC, which provides a promising prospect for the treatment of CRC.
Herein, we aimed to explore the role of circRNAs which are strongly correlated with the carcinogenesis of CRC. A novel circRNA hsa_circ_102049 (circTADA2A) was found to be the significantly downregulated in CRC. Function studies were carried out to investigate the role of circTADA2A on CRC progression. Mechanistically, we predicted and verified the miRNA sponged by circTADA2A. We elucidated the regulatory function and molecular mechanism of circTADA2A in the initiation and evolution of CRC.
Materials and Methods
Tissue Sample Collection and Cell Culture
A total of 25 tumor samples and paired non-cancer tissues were acquired from CRC patients diagnosed in Ningbo No.1 Hospital. All the participants signed written informed consent. Fresh tissues detached from patients were immediately treated with liquid nitrogen and subsequently preserved at −80°C for RNA extraction. This study was approved by the Ethics Review Board of Ningbo No.1 Hospital. All the procedures were complied with the guidelines of the Declaration of Helsinki.
Five human CRC cell lines (LOVO, SW620, SW480 and HT-29) and normal human colorectal epithelial cell line FHC were all supplied by American Type Culture Collection (ATCC, Manassas, USA). All the cells were maintained in DMEM (Invitrogen, Carlsbad, USA) or RPMI-1640 (Invitrogen) added with 10% FBS, 100 mg/L streptomycins as well as 100 mg/L penicillins under a 37°C moist atmosphere containing 5% CO2.
Cell Transfection
Short hairpin RNAs (shRNAs) against circTADA2A via targeting back-splice junction were procured from GeneChem (Shanghai, China). Non-specific shRNAs were utilized as a negative control. For upregulation of circTADA2A, circTADA2A overexpressing plasmid and negative control empty vector were designed and synthesized by GeneChem. The mimic and inhibitor of miR-1229 inhibitor, miRNA mimic negative control (miR-NC) and miRNA inhibitor negative control (inhibitor-NC) were acquired from RiboBio (Guangzhou, China). To knock down BCL2L10 expression, small interfering RNA for BCL2L10 (siBCL2L10) and scrambled siRNA were purchased from GeneChem. Cell transfection was performed with Lipofectamine 2000 (Invitrogen, USA) following the manufacturer’s protocols.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA from clinical tissues and cells was extracted with TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Thereafter, cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Dalian, China) and Mir-X miR First-Strand Synthesis Kit (TaKaRa, Dalian, China). qRT-PCR was implemented on 7500FAST Real-time PCR System (ABI, USA) using a SYBR Green PCR kit (Takara, Dalian, China) in line with the vendor’s directions. GAPDH and U6 served as inherent controls for normalization. The gene expression level was calculated with the 2−ΔΔCt method. Each sample was analyzed three times.
RNase R and Actinomycin D Treatment
To verify the features of circRNA, 10 μg of total RNA was treated with 40U RNase R for 1 h at 37°C. For estimation of stability, the medium was complemented with 2 mg/mL Actinomycin D or negative control dimethyl sulfoxide (DMSO). Following treatment, the expression of circTADA2A and linear TADA2A was subjected to qRT-PCR analysis.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK8) assay was conducted for evaluation of cell proliferation. Following transfection, LOVO and HT-29 cells were inoculated into 96-well plates at a density of 1000 cells per well and cultivated at 37°C. Each well was supplemented with 10 μL of CCK 8 solution (Dojindo, Kumamoto, Japan) at different time points (0 h, 24h, 48 h and 72h) after incubation. Then, cells were cultured at 37°C for another 2 h and the optical density (OD) value was examined at 490nm with a microplate reader.
For colony formation assay, transfected LOVO and HT-29 cells were seeded in 6-well plates and cultured at 37°C for 2 weeks. The culture medium was replaced every three days. After washing with PBS, the colonies were immobilized using 4% paraformaldehyde, stained by 0.1% crystal violet and photographed by a microscope.
Transwell Migration Assay
Cell migration was detected by transwell migration assay using transwell chambers (Corning, Cambridge, USA) obeying the instructions of the manufacturer. In short, cell suspension and 200 μL serum-free medium were placed in the upper chambers, and the bottom of chambers was complemented with 600 μL complete medium containing 20% FBS. At 24 h post-incubation, migrated cells were fixed by 4% paraformaldehyde and treated with 0.5% crystal violet. The images of migrated cells were obtained with a microscope in five randomly selected fields.
Western Blot
Transfected LOVO and HT-29 cells were lysed by using lysis buffer supplemented with protease inhibitors. 40 μg amounts of protein extracts were electrophoresed on 10% SDS-PAGE and transferred to PVDF membranes (Millipore, USA). After blocking in 5% skimmed milk for 2 h, membranes went through an overnight incubation with primary antibody against BCL2L10 at 4°C. Then, PVDF membranes were probed by the appropriate secondary antibody for 2 h at room temperature and examined with an enhanced chemiluminescent kit (Millipore, USA). GAPDH was employed as the endogenous reference.
RNA Pull-Down Assay
RNA pull-down assay was carried out with the Magnetic RNA-Protein Pull-Down Kit (Pierce, MA, USA). Briefly, transfected cells were harvested, followed by trypsinization, sonicated and incubated with biotinylated miR-NC or miR-1229 and streptavidin magnetic beads overnight at 4°C. The expression of circTADA2A and BCL2L10 in eluted RNAs was determined by qRT-PCR.
Luciferase Reporter Gene Assay
The sequences of circTADA2A and BCL2L10 3’UTR containing miR-1229 binding sites were inserted into luciferase vectors pmirGLO (Promega, USA) to construct wild-type circTADA2A and BCL2L10 plasmids. The mutant vectors were acquired by mutating binding sites with miR-1229. Afterward, LOVO and HT-29 cells were co-transfected with wild-type or mutant plasmids and miR-1229 mimic or miR-NC by utilizing Lipofectamine 2000 (Invitrogen, USA) following the instructions recommended by the vendor. 48 h later, luciferase activity was tested with the Dual-Luciferase Reporter Assay System (Promega, USA).
Statistical Analysis
The experimental data were analyzed with SPSS software 20.0 (IBM, Armonk, USA). All the results were presented as the mean ± SEM and each assay was replicated at least three times. Comparisons between two or more groups were conducted using Student’s t-test and ANOVA. Pearson correlation analysis was applied to estimate the correlation among circTADA2A, miR-1229 and BCL2L10. Differences were considered to indicate statistically significant when P < 0.05.
Results
circTADA2A Was Lowly Expressed in CRC
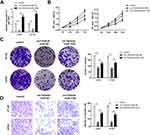
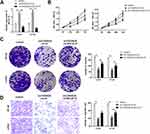
CircRNA is stable due to the ring structure. Thus, we verified the stability of the novel circRNA (circTADA2A) in CRC cells. The results indicated that circTADA2A was more stable than its linear isoform in response to RNase R digestion (Figure 1A). In agreement with the above findings, our observations revealed that circTADA2A was more resistant to actinomycin D (ActD) treatment in contrast to its linear mRNA, further validating the circular characteristics of circTADA2A (Figure 1B). To confirm the expression pattern of circTADA2A in CRC, qRT-PCR was conducted in the tissues and cell lines of CRC. Compared to the adjacent normal tissue samples, circTADA2A expression was markedly diminished in tumor specimens of CRC patients (Figure 1C). Similarly, the expression level of circTADA2A was lower in five CRC cell lines than that in normal colorectal cell line (Figure 1D). Taken together, circTADA2A was downregulated in CRC.
circTADA2A Led to the Suppression of Cell Proliferation and Migration in CRC
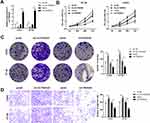
To identify the functional role of circTADA2A in CRC, we knocked down circTADA2A expression in LOVO and HT-29 cells by transfection with shRNA targeting circTADA2A (Figure 2A). Besides, for ectopic expression of circTADA2A, circTADA2A overexpressing vectors were successfully transfected into LOVO and HT-29 cells and then qRT-PCR analysis certified the overexpression efficiency (Figure 2A). The CCK-8 assay showed that silencing circTADA2A prominently promoted the viability of LOVO and HT-29 cells, overexpression of circTADA2A inhibited cell proliferation (Figure 2B). Additionally, depletion of circTADA2A remarkably restrained the colony-forming capability of LOVO and HT-29 cells. On the contrary, upregulation of circTADA2A resulted in the opposite consequence (Figure 2C). The transwell assay demonstrated that circTADA2A knockdown augmented the number of migrated cells, while LOVO and HT-29 cells overexpressing circTADA2A displayed the reduced capacity for migration (Figure 2D). By the large, these findings provided strong evidence that circTADA2A played a tumor suppressor role in CRC.
circTADA2A Functioned as a miRNA Sponge for miR-1229
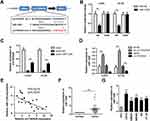
In view of the dominating location of circTADA2A in the cytoplasm, we therewith investigated miRNAs targeting circTADA2A through the competitive endogenous (ceRNA) mechanism. With the aid of bioinformatics analysis, miR-1229 was found to own the potential to bind with circTADA2A (Figure 3A). As shown in Figure 3B, miR-1229 mimic obviously lowered the luciferase activity of wild-type circTADA2A as opposed to miR-NC, whereas no significant change occurred in the luciferase activity of mutated circTADA2A (Figure 3B). Consistently, RNA pull-down assay delineated that circTADA2A was abundantly expressed in mixture pulled down by miR-1229, confirming the binding relationship between circTADA2A and miR-1229 (Figure 3C). Furthermore, we observed that downregulation of circTADA2A caused the increased level of miR-1229 and circTADA2A overexpression weakened miR-1229 expression (Figure 3D). Results of Pearson correlation analysis illuminated the inverse association of circTADA2A expression with miR-1229 level in clinical tissue samples (Figure 3E). In addition, qRT-PCR assay revealed that miR-1229 was expressed at a higher level in CRC tissues and cells compared to normal tissue samples and cells (Figure 3F and G). Based on the foregoing results, we concluded that miR-1229 was sponged by circTADA2A.
Ectopic Expression of miR-1229 Abolished the Regulatory Function of circTADA2A in CRC
In order to explore whether circTADA2A exerted its anti-tumor property in CRC by modulation of miR-1229, we carried out rescue experiments. qRT-PCR assay suggested that the declined expression of miR-1229 caused by circTADA2A overexpression was recovered when LOVO and HT-29 cells were transfected with miR-1229 mimic (Figure 4A). Results of CCK-8 assay exhibited that miR-1229 mimic reversed the circTADA2A-mediated inhibition of cell proliferation in CRC (Figure 4B). In concert with these findings, the colony formation assay further validated the role of circTADA2A/miR-1229 in the proliferative capacity of LOVO and HT-29 cells (Figure 4C). Furthermore, it was disclosed by transwell assay that overexpression of miR-1229 reinstated the migration of LOVO and HT-29 cells suppressed by ectopic expression of circTADA2A (Figure 4D). In a word, these results supported the hypothesis that miR-1229 was responsible for the effects of circTADA2A on cell proliferation and migration in CRC.
circTADA2A Protected BCL2L10 from miR-1229-Mediated Suppression via Acting as a ceRNA
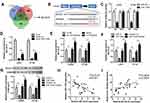
Given that miRNAs are participated in tumor progression by regulating target genes at post-transcriptional level, we sought to screen candidate targets of miR-1229 using 3 bioinformatics tools (Targetscan, miRDB and DIANA) and uncovered that BCL2L10 was a potential functional target of miR-1229 (Figure 5A and B). Afterwards, luciferase reporter assay manifested that miR-1229 mimic only lessened the luciferase activity of wild-type BCL2L10, but did not alter the activity of the mutant form (Figure 5C). Similarly, RNA pull down assay justified that miR-1229 directly bound to BCL2L10 (Figure 5D). Considering the ceRNA theory, we determined the impacts of circTADA2A on BCL2L10 expression. Our data showed that depletion of circTADA2A triggered the decrease of BCL2L10 level, overexpression of circTADA2A enhanced BCL2L10 expression (Figure 5E). Inversely, miR-1229 mimic reduced the expression of BCL2L10, and suppression of miR-1229 led to the opposite result (Figure 5F). Western blot further demonstrated the inhibitory role of miR-1229 in BCL2L10 protein expression level (Figure 5G). Of note, the negative association between miR-1229 and BCL2L10 expression as well as the positive relationship between circTADA2A and BCL2L10 expression were obtained in tissue specimens, as evidenced by a Pearson correlation analysis (Figure 5H and I).
circTADA2A Inhibited the Malignant Behavior of CRC Cells by Regulating miR-1229/BCL2L10 Axis
Finally, we intended to confirm whether the suppressive influences of circTADA2A on CRC cell proliferation and migration were mediated by BCL2L10. Hence, LOVO and HT-29 cells were treated with si-BCL2L10 following transfection with circTADA2A overexpressing plasmids. Results of qRT-PCR analysis suggested that ectopic expression of circTADA2A contributed to the overt increase of BCL2L10 level and the recovery of BCL2L10 expression occurred due to silencing of BCL2L10 (Figure 6A). The CCK-8 and colony formation assays certified that cell proliferation repressed by circTADA2A overexpression was retrieved in response to depletion of BCL2L10 (Figure 6B and C). Furthermore, we observed that knockdown of BCL2L10 abrogated the anti-tumor effects of circTADA2A on the migratory ability of LOVO and HT-29 cells (Figure 6D). Collectively, the above findings manifested that circTADA2A impeded CRC cell proliferation and migration via targeting miR-1229-mediated BCL2L10.
Discussion
Colorectal cancer is recognized as one of the most frequently diagnosed malignancies in digestive system, resulting in a myriad of people dying from this disease worldwide.19 To a certain extent, progress has been obtained in clinical interventions for CRC, contributing to the prolongation of patients’ life expectancy.20 However, the prognosis of patients with CRC remains far from favorable on account of delayed diagnosis, metastasis, radioresistance and recurrence.21,22 Hence, it is necessary to explore the pathogenesis of CRC and seek for diagnostic and therapeutic targets in order to ameliorate the curative efficacy of CRC.
With great advances in molecular biology technology, various lines of evidence highlight that circRNAs are involved in numerous cell activities of CRCr.23,24 For instance, circHIPK3 contributes to cell growth and metastasis of CRC via sponging miR-7.25 Exosome-delivered hsa_circ_0005963 facilitates glycolysis to enhance chemoresistance in CRC by the miR-122-PKM2 axis.26 CircRNA_101951 accelerates CRC cell migration and invasion through activating the KIF3A-mediated EMT pathway.27 The results of microarray analysis unveiled that the differential expression of circTADA2A was the most significant in CRC tissues among down-regulated circRNAs. Interestingly, circTADA2A has been testified to exert opposite roles in osteosarcoma and breast cancer.28,29 As in CRC, circTADA2A has been reported to play a anti-cancer role via miR-374a-3p/KLF14 axis.30 Nonetheless, the potential of circTADA2A and its latent mechanism in CRC remain unclear. Hence, we focused on characterizing the functional role of circTADA2A in CRC development. Our findings revealed that the expression of circTADA2A was lower in CRC specimens and cells than that in normal tissue samples and cells. Here, the results are based on a small number of patients. This is a limitation of the present study. We will verify these findings on a large number of patients in our further study. Moreover, functional assays manifested that circTADA2A played a tumor suppressor role in the malignant behaviors of CRC cells. However, animal study has not been carried out to further confirm the anti-cancer role of circTADA2A in vivo, which is another limitation of this study. Here, we will conduct the in vivo study and explore the role of circTADA2A in near future.
The ceRNA hypothesis emphasizes that transcripts containing miRNA-binding sites compete with each other in the regulatory network at the post-transcriptional level.31 Accumulating investigations support the above hypothesis and prove that circRNAs modulate the activity of miRNAs to affect the expression of target genes by serving as miRNA sponges.32,33 FISH assay showed that circTADA2A was principally localized in the cytoplasm of CRC cells, unraveling that circTADA2A might execute its performance in cell proliferation and migration of CRC through a ceRNA-dependent mechanism. Through browsing circinteractome database, circTADA2A was discovered to harbor the predicted binding sites with miR-1229. Of note, previous researches have demonstrated that upregulation of miR-1229 is a prognostic indicator for CRC.34,35 Thus, we selected miR-1229 for in-depth study. Subsequently, our findings confirmed that circTADA2A exhibited the strong-binding capacity for miR-1229 and negatively regulated miR-1229 expression. More importantly, the anti-tumor property of circTADA2A in CRC was mediated by miR-1229. Moreover, BCL2L10, which has been validated to be a tumor suppressor gene in ovarian cancer, gastric cancer and hepatocellular carcinoma,36–38 was found to be a potential functional target gene of miR-1229 via bioinformatics analysis. Further assay demonstrated that BCL2L10 directly bound to miR-1229 and circTADA2A affected BCL2L10 expression level through competing for miR-1229. Finally, we unveiled that suppression of BCL2L10 abolished the inhibitory impacts of circTADA2A on CRC progression.
To the best of our knowledge, the present study shed light on the function and molecular mechanism of circTADA2A in CRC for the first time. To sum up, our results certified that circTADA2A restrained the proliferation and migration of CRC cells by sponging miR-1229 to modulate BCL2L10, which provided experimental evidence for the possibility of circTADA2A as a novel therapeutic biomarker for CRC.
Acknowledgments
The authors appreciate the support from Ningbo No.1 Hospital.
Funding
This study was funded by the Traditional Chinese Medicine Science and Technology Project of Zhejiang Province (grant no. 2018ZA109); Zhejiang Science and Technology Department Fund (grant no.2019KY566; Ningbo Public Welfare Science and Technology Project (grant no.202002N3163).
Disclosure
The authors declare that there is no conflict of interest in this paper.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi:10.1016/S0140-6736(13)61649-9
3. Yang L, Parkin DM, Li LD, et al. Estimation and projection of the national profile of cancer mortality in China: 1991–2005. Br J Cancer. 2004;90(11):2157–2166. doi:10.1038/sj.bjc.6601813
4. Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17(4):268. doi:10.1038/nrc.2017.24
5. Smith RA, von Eschenbach AC, Wender R, et al. American Cancer Society guidelines for the early detection of cancer: update of early detection guidelines for prostate, colorectal, and endometrial cancers. Also: update 2001–testing for early lung cancer detection. CA Cancer J Clin. 2001;51(1):38–75. doi:10.3322/canjclin.51.1.38
6. Yazgan O, Krebs JE. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem Cell Biol. 2007;85(4):484–496. doi:10.1139/O07-061
7. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi:10.1038/nrd.2016.117
8. Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. doi:10.1002/path.2638
9. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
10. Conn SJ, Pillman K, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi:10.1016/j.cell.2015.02.014
11. Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi:10.1186/s13059-014-0409-z
12. Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
13. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
14. Ju H, Zhao Q, Wang F, et al. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol Med. 2019;11(10):e10168. doi:10.15252/emmm.201810168
15. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. doi:10.1242/dev.128074
16. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
17. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
18. Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi:10.1038/cdd.2017.86
19. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
20. Jin Y, Yu LL, Zhang B, et al. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51(12):e7811. doi:10.1590/1414-431x20187811
21. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
22. Cantero-Muñoz P, Urién MA, Ruano-Ravina A. Efficacy and safety of intraoperative radiotherapy in colorectal cancer: a systematic review. Cancer Lett. 2011;306(2):121–133. doi:10.1016/j.canlet.2011.02.020
23. Xu H, Wang C, Song H, et al. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. doi:10.1186/s12943-018-0932-8
24. Chen S, Zhang L, Su Y, et al. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39(6):2499–2512.
25. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
26. Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi:10.1002/1878-0261.12629
27. Li YF, Pei FL, Cao MZ. CircRNA_101951 promotes migration and invasion of colorectal cancer cells by regulating the KIF3A-mediated EMT pathway. Exp Ther Med. 2020;19(5):3355–3361.
28. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi:10.1186/s12943-019-1007-1
29. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. doi:10.1038/s41419-019-1382-y
30. Li Z, Yao H, Wang S, et al. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res. 2020;39(1):1–14.
31. Salmena L, L. P, Y. T, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
32. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
33. Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi:10.1038/nature09144
34. Song W, Fu T. Circular RNA-associated competing endogenous RNA network and prognostic nomogram for patients with colorectal cancer. Front Oncol. 2019;9:1181. doi:10.3389/fonc.2019.01181
35. Hu HY, Yu CH, Zhang HH, et al. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470–477. doi:10.1016/j.ijbiomac.2019.03.221
36. Lee SY, Kwon J, Woo JH, et al. Bcl2l10 mediates the proliferation, invasion and migration of ovarian cancer cells. Int J Oncol. 2020;56(2):618–629.
37. Xu J, Cao X, Long Z, et al. BCL2L10 protein regulates apoptosis/proliferation through differential pathways in gastric cancer cells. J Pathol. 2011;223(3):400–409. doi:10.1002/path.2811
38. Bai Y, Wang J, Han J, et al. BCL2L10 inhibits growth and metastasis of hepatocellular carcinoma both in vitro and in vivo. Mol Carcinog. 2017;56(3):1137–1149. doi:10.1002/mc.22580
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.