Back to Journals » Cancer Management and Research » Volume 13
CircSND1 Regulated by TNF-α Promotes the Migration and Invasion of Cervical Cancer Cells
Authors Bai L, Sun W , Han Z, Tang H
Received 27 October 2020
Accepted for publication 9 December 2020
Published 12 January 2021 Volume 2021:13 Pages 259—275
DOI https://doi.org/10.2147/CMAR.S289032
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Chien-Feng Li
Lili Bai,* Wangjie Sun,* Zhe Han, Hua Tang
Tianjin Life Science Research Center, Tianjin Key Laboratory of Inflammation Biology, Collaborative Innovation Center of Tianjin for Medical Epigenetics, Department of Pathogen Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hua Tang
Tianjin Life Science Research Center, Tianjin Key Laboratory of Inflammation Biology, Collaborative Innovation Center of Tianjin for Medical Epigenetics, Department of Pathogen Biology, School of Basic Medical Sciences, Tianjin Medical University, 22 Qi-Xiang-Tai Road, Tianjin 300070, People’s Republic of China
Tel/Fax +86 22 23542503
Email [email protected]
Aim: To explore the role and potential mechanism of circSND1 in cervical cancer (CC).
Main Methods: qRT-PCR was used to determine the expression of circSND1 in tumor necrosis factor-α (TNF-α)-treated HeLa cells. CircSND1 overexpression and knockdown were performed to indicate the functional role of circSND1 in vitro and in vivo. Luciferase assay was used to analyze promoter activity. The expression and regulation of circSND1, miR-125a-3p and FUT6 were evaluated using EGFP fluorescent reporter assay and rescue experiments. Immunofluorescence and Western blot assays were used to analyze the activation of nuclear factor-κB (NF-κB).
Results: In HeLa cells, TNF-α up-regulated the expression of circSND1 by activating the NF-κB signaling pathway. Overexpression of circSND1 significantly increased the migration and invasion and the epithelial–mesenchymal transition (EMT) process of CC cells, and promoted tumor metastasis in xenograft nude mouse model, whereas down-regulation of circSND1 exerted opposite effects. Furthermore, circSND1 enhanced the expression of FUT6 via sponging miR-125a-3p, and FUT6 activated NF-κB signaling pathway.
Conclusion: We found that circSND1 promoted the expression of FUT6 and the malignant behavior of cervical cancer through the ceRNA mechanism, and there was a TNF-α/NF-κB/circSND1/miR-125a-3p/FUT6/NF-κB positive feedback pathway between them, which suggests that circSND1 can be a promising prognostic marker and therapeutical target for cervical cancer.
Keywords: circSND1, miR-125a-3p, FUT6, NF-κB, ceRNA, cervical cancer
Introduction
Cervical cancer (CC) is one of the most common gynecological cancer. Over 100 genotypes of human papillomavirus (HPV) have been identified and persistent infection by certain high-risk HPV types has been recognized as a major cause of cervical cancer.1 In recent years, cytology combined with HPV screening programs has been applied successfully in cervical cancer treatment and proved to be an effective strategy to decrease the morbidity of cervical cancer.2 However, the prognosis and the 5-year survival rates of CC patients remain unsatisfying.3 Therefore, further exploration of the mechanism of cervical cancer occurrence and development is imperative for disease diagnosis, treatment and prognosis.
Circular RNAs (circRNAs) are a novel class of non-coding RNAs, which form a covalently closed loop structure without 5′ cap and 3′ poly(A) tail.4 In 1976, Sanger et al first discovered RNA in a circular form in a viroid.5 In 1979, Hsu et al used electron microscopy for observation and discovered that there is also a circular conformation of RNA in the cytoplasm of HeLa cells.6 Initially, circRNA was considered as a result of abnormal splicing of RNA and attracted little attention. In recent years, more and more circRNAs were observed by high-throughput sequencing techniques.7 CircRNAs have been found as tissue-specific and played vital roles in the occurrence and progression of cancer.8,9 Accumulated evidences showed that circRNAs function as miRNA sponges through the competing endogenous RNA (ceRNA) network.10 It was reported that circRNA cSMARCA5 inhibits proliferation and metastasis by sponging miR-17-3p and miR-181b-5p to promote the expression of TIMP3 in hepatocellular carcinoma cells while cTFRC facilitates progression of bladder cancer by sponging miR-107.11,12 However, the exact role and mechanism of circRNAs in CC remain elusive to date.
Unresolvable inflammation is closely linked with the initiation and development of malignant tumors. In the inflammatory environment, the main pro-inflammatory factors (TNF-α, IL-6 and others) directly or indirectly activate the downstream NF-κB signal pathway and regulate the expression of a series of oncogenes and tumor suppressor genes, including a large amount of non-coding RNAs.13 A large number of circRNAs have important regulatory effects on the NF-κB signaling pathway. For example, Guan et al found that circPUM1 promotes the expression of NF-κB by adsorbing miR-615-5p, thereby promoting the occurrence and development of ovarian cancer.14 In addition, circular RNA circC3P1 inhibits kidney cancer cell activity by regulating miR-21/PTEN axis and inactivating PI3K/AKT and NF-κB pathways.15 However, the relationship between inflammation and cervical cancer is still unclear. The purpose of our research is to explore whether dysregulated circRNA plays a role in it.
In this study, we used next-generation sequencing of circRNAs to screen the differentially expressed circRNAs in TNF-α-induced HeLa cells and identified the significantly upregulated circSND1, which is derived from the exons 8 to 10 of the SND1 gene. We demonstrated here that TNF-α notably induced circSND1 expression by transcription factor NF-κB, which directly bound to the promoter region of circSND1. CircSND1 promoted metastasis of CC by acting as a “sponge” of miR-125a-3p to upregulate the expression of FUT6, which subsequently activated NF-κB signaling pathway. Therefore, circSND1 exerted its functions via the TNF-α/NF-κB/circSND1/miR-125a-3p/FUT6/NF-κB positive regulatory circuit. This is the first report on the expression and role of circSND1 in CC. Our findings indicated that circSND1 may act as a promising diagnostic and prognostic marker for CC.
Materials and Methods
Cell Culture and Transfection
Human cervical cancer cell lines HeLa and SiHa were supplied by the American Type Culture Collection (ATCC) cell bank (HeLa, ATCC® CCL-2™. SiHa, ATCC® HTB-35™). The cells were cultured in RPMI 1640 (HeLa) and DMEM (SiHa) medium (GIBCO BRL, Gaithersburg, MD) respectively. The complete culture medium contained 7% (HeLa) or 10% (SiHa) fetal bovine serum, 100 ug/mL streptomycin and 100 IU/mL penicillin. All cells were cultured in 5% CO2 at 37°C. All transfection experiments were carried out by using LipofectamineTM 2000 transfection Reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instruction.
Plasmid Construction
A fragment containing the sequence of circSND1 was amplified and cloned into the pcDNA3-vector as an over-expression plasmid (pcDNA3-vector/circSND1). The pcDNA3-vector was constructed following the report.16 The full CDS sequence of FUT6 and p65 was amplified and cloned into pcDNA3 (pcDNA3-flag/FUT6 and pcDNA3-flag/p65) and pSilencer 2.1-U6 neo was used to construct the shRNA vectors to silence circSND1 and FUT6 expressions. The synthesized oligonucleotides were annealed and ligated into pSilencer. The fragment containing miR-125a-3p binding sites in circSND1 was synthesized, annealed and ligated into pcDNA3-EGFP to construct the reporter vectors (pcDNA3-EGFP/circSND1-3’UTR-WT/Mut). The over-expression plasmids (pcDNA3/miR-125a-3p, pcDNA3-EGFP/FUT6-3’UTR-WT/Mut), antisense oligonucleotides to silence miR-125a-3p and the respective control (ASO-miR-125a-3p, ASO-NC) were commercially synthesized (GenePharma, Shanghai, China). The fragments containing sequence of circSND1 promoter were cloned into pGL3-Basic vector. All sequences of primers and oligonucleotides were listed in (Table 1).
 |  |  |
Table 1 Primers and Oligonucleotides Used in This Study |
RNA Extraction and RT-qPCR
TRIzol reagent (Invitrogen, Carlsbad, CA) was adopted to extract total RNAs. A Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) was used to measure RNA concentration. RNAs were reverse transcribed into cDNA by using M-MLV (Promega, Madison, WI). For examination of circSND1, the cDNA was synthesized with random primers rather than oligo(dT). And SYBR Premix Ex TaqTM (TaKaRa, Dalian, China) was adopted in RT-qPCR experiments. β-actin and U6 were used as the internal control genes for mRNA and miRNA, respectively. The relative fold-change was calculated by the 2−ΔΔCt method. All primer sequences for RT-qPCR were provided in (Table 1).
Nuclear-Cytoplasmic Fractionation, RNase R Actinomycin D Treatment and BAY-11-7082
RNAs from nucleus and cytoplasm of CC cells were separated by the RNA Subcellular Isolation Kit (Active Motif, USA) following the manufacturer’s instructions. RNase R treatment was executed at 37°C with 4 U/μg of RNase R (Epicenter Biotechnologies, Madison, WI, USA) for 30 min. Total RNA from HeLa cell was treated with 5 μg/mL actinomycin D (Sigma, USA) against new RNA synthesis for 2 h,4 h,8 h,12 h and 24 h, respectively. An NF-κB inhibitor BAY-11-7082 (MCE, USA) was used to treat HeLa cells at 25μM with 1h.
Transwell Migration and Invasion Assays, Colony Formation and MTT Assay
Migration and invasion assays were performed as previously described.17,18 In brief, for migration and invasion, each well was photographed with a microscope and the number of cells in five fields was counted and the average number was taken as the number of cells that migrated and invaded. For clone formation experiments, a single cell cluster with a cell count more than 50 under the microscope was considered a clone. Clone formation rate = the number of clones/the number of cells inoculated. For MTT assay, relative cell viability = absorbance of experimental group/absorbance of control group. Normalize the control group to 1 in data statistics.
Immunofluorescence
The subsequent protocol was previously reported by Chen et al.19 Specific primary antibodies against FUT6 (anti‐rabbit, diluted 1:20) was obtained from Tianjin Saier Biotech (Tianjin, China), and Flag-p65 (anti‐mouse, diluted 1:50) was obtained from Sigma (United States). Fluorescent-conjugated secondary antibodies TRITC and FITC were obtained from ABclonal (United States). DAPI was obtained from Beyotime (Shanghai, China). Images were captured by confocal microscopy.
Western Blot Analysis
Western blot analysis was performed as previously described.20 The antibodies were used as follows: EGFP (Saier Biotechnology, SRP12235; 1:1000), FUT6 (Saier Biotechnology, SRP06891; 1:500), p65 (Saier Biotechnology, SRP00533; 1:500), E-cadherin (Saier Biotechnology, SRP05266; 1:100), vimentin (Saier Biotechnology, SRP01327; 1:1000) HPV-18-E6 (Saier Biotechnology, LXR28/CH; 1:500), HPV-18-E7 (Saier Biotechnology, LXR29/CH; 1:500), HPV-16-E6 (Saier Biotechnology, LXR23/DR; 1:500), HPV-16-E7 (Saier Biotechnology, LXR24/CH; 1:500), CENPA (Saier Biotechnology, SRP01348; 1:10,000) and GAPDH (Saier Biotechnology, SRP00849; 1:5000). The secondary goat anti-rabbit antibody was obtained from Sigma (Sigma, R0881). Whole cell lysate and cytoplasmic proteins were normalized with an internal reference protein GAPDH, while nuclear proteins were normalized with CENPA. And the expression intensities of all other proteins were standardized with an internal reference protein before the comparison between the experimental group and the control group. The band intensities were quantified by ImageJ.
Immunohistochemistry
The tumors separated from the nude mice were prepared for paraffin sections. Paraffin sections were dewaxed with xylene and gradient alcohol, rinsed with three distilled water and placed in 98°C sodium citrate buffer for 10–15 minutes for antigen thermal restoration. Then, wash the sections with PBS, and incubate with 3% H2O2 at 37°C for 15 minutes to block endogenous peroxidase activity. Blocked with 5% BSA for 1 hour, added primary antibody against FUT6 (Saier Biotechnology, SRP06891; 1:500), and incubated overnight at 4°C. Rinse the sections with PBS, add biotinylated two holes, and at 37°C for 40 minutes, developed with DAB chromogenic reagent and observe under a microscope, evaluate the staining result by counting the percentage of positive cells.
Animal Experiments
Six-week-old female BALB/c nude mice were selected for in vivo xenotransplantation experiments. The mice were kept in SPF environment. All procedures were approved by the Animal Protection and Use Committee of Tianjin Medical University. The HeLa cell line stably expressing circSND1 and the empty vector was constructed and 5×105 cells were injected into the tail vein. After 60 days, the mice were sacrificed to isolate their lungs. Lung metastatic nodules were counted and assessed by hematoxylin and eosin (HE) staining. The expression levels of RNA and protein from Lung tissue were determined with the above method.
Statistical Analysis
All experimental data were processed with GraphPad Prism 8.0 software. The data are presented as the means ± SD. The two-tailed Student’s t-test was applied for contrast between the two groups. ANOVA analysis was performed for the analysis of MTT assay. P < 0.05 was considered as statistically significant (*p<0.05; **p<0.01; ***p<0.001). The results from each set of experiments were repeated at least three times.
Results
CircSND1 is Up-Regulated by TNF-α in CC Cells
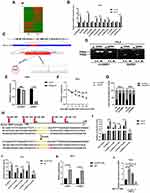
To study the regulatory relationship between circRNAs and inflammation and cervical cancer, we performed circRNAs expression profiles in TNF-α-treated-HeLa cells by high-throughput sequencing. Compared with the non-treated group, 98 circRNAs were up-regulated and 63 circRNAs were down-regulated (Figure 1A). By further analysis of the read counts and the FPKM value, 11 most up-regulated circRNAs were chosen for further validation by RT-qPCR. The results indicated that the expression levels of 6/11 circRNAs were increased greatly after TNF-α treatment (Figure 1B). Among them, circSND1 (hsa_circ_0003655) showed the most significant upregulation, thus was chosen for further study.
CircSND1 (hsa_circ_0003655) was generated from exon 8, exon 9 and exon10 of the SND1 gene by back-splicing (312 bp) based on the annotation of circBase (http://www.circbase.org/). Its back-splicing junction was confirmed by Sanger sequencing (Figure 1C). In order to eliminate the possibility of genomic rearrangements and trans-splicing, PCR and agarose gel electrophoresis assays were performed to detect the expression level of backspliced or canonical forms of SND1 in cDNA and genomic DNA (gDNA). Divergent primers detected circSND1 in cDNA, but no products were detected in gDNA (Figure 1D). In addition, circSND1 was found as more resistant to RNase R digestion than mSND1 (Figure 1E). Actinomycin D was used to inhibit the RNA synthesis, circSND1 showed high stability after treatment by actinomycin D compared with mSND1 (Figure 1F). Then, nuclear-cytoplasmic fractionation assays showed that circSND1 was mainly located in the cytoplasm of CC cells (Figure 1G). To investigate the mechanism by which TNF-α induced circSND1 expression, bioinformatics analysis was used to predict the transcription factor bound to the SND1 promoter. We found that circSND1 promoter region contains three potential NF-κB binding sites which is a target gene of TNF-α.21,22 To study whether TNF-α promotes the expression of circSND1 via NF-κB, NF-κB binding sites (at −116 mut#1, −174 mut#2 and −267 mut#3) in promoter sequences of SND1 were selected and luciferase reporter plasmids containing the wild type (WT) or mutant (mut) SND1 promoter were constructed (Figure 1H). According to results from luciferase reporter assays, TNF-α promoted the activity of wild type SND1 promoter and partially promoted the activities of mut#1 and mut#2 promoter; while TNF-α had no obvious effect on the activities of mut#3 promoter. When the two binding sites were simultaneously mutated (at −116 and −174), the promotion of TNF-α was abolished (Figure 1I). After this, we assessed the induction of p65 on circSND1 promoter activity and found similar results with overexpressing of p65 (Figure 1J). RT-qPCR assay demonstrated that overexpression of p65 increased the mRNA level of circSND1 and mSND1 (Figure 1K). Subsequently, to further confirm that TNF-α promotes the expression of circSND1 through NF-κB, we studied the effect of TNF-α on the expression of circSND1 in the pretreatment of the NF-κB inhibitor BAY 11–7082. RT-qPCR assays showed that compared with the control without BAY 11–7082, the expression level of circSND1 was significantly reduced after HeLa cells were treated with BAY 11–7082. Meanwhile, the promotion effect of TNF-α on circSND1 was significantly weakened after treating HeLa with BAY 11–7082 (Figure 1L). These results showed that the up-expression of circSND1 was induced by TNF-α through activation of the NF-κB signal pathway in HeLa cells.
CircSND1 Enhances Migration and Invasion of CC Cells in vitro
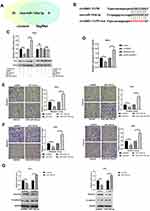
To determine the effects of circSND1 on cervical cancer cells, circSND1 overexpression and knockdown plasmids were constructed and the effectiveness of the plasmids was verified by RT-qPCR (Figure 2A). Since pshR-circSND1#2 was more effective, it was adopted in the further experiment and referred to as pshR-circSND1. Then, functional experiments were performed in HeLa and SiHa cells. As shown in (Figure 2B and C), overexpression of circSND1 accelerated, whereas knockdown of circSND1 significantly attenuated cell migration and invasion abilities. However, overexpression of circSND1 did not affect cell viability (Figure 2D) or colony formation ability in CC cells (Figure 2E). To further investigate the potential mechanisms of how circSND1 promotes the migration and invasion ability of CC cells, we detected the expression of key EMT markers, E-cadherin and Vimentin. Western blot analysis revealed that overexpression of circSND1 led to a reduction of E-cadherin expression, which is an epithelial marker, but it elevated the expression of mesenchymal markers such as Vimentin (Figure 2F). Among the high-risk types, HPV 16 is undoubtedly the most frequently detected genotype in 60.5% of cervical cancer cases, followed by HPV18. Oncoproteins encoded by E6 and E7 genes are important factors that cause cervical epithelial cancer.23,24 To determine whether circSND1 has an effect on the two oncoproteins of HPV in the process of CC, we detected the expression levels of E6/E7 oncogenes after overexpression of circSND1 in HeLa (harboring HPV 18 type) and SiHa (harboring HPV16 type) cells with Western blot assay. Results showed that circSND1 had no obvious effect on the expression level of E6 and E7 (Figure 2G). These data indicated that circSND1 facilitated the migration and invasion of CC cells and EMT process.
CircSND1 Serves as miR-125-3p Sponge in CC Cells
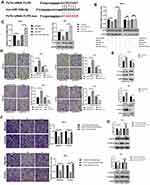
We know that circRNAs can act as a miRNA sponge to regulate gene expression. In order to determine whether circSND1 can function by ceRNA mechanism, we first predicted the miRNAs that might bind to circSND1 through the circbank database (http://www.circbank.cn/) and the RegRNA2.0 database (http://www.circbank.cn/) (Figure 3A). Combined with previous literature reports,25,26 miR-125a-3p was chosen for further study. The binding sites for miR-125a-3p in circSND1 sequence were shown in (Figure 3B.) To explore whether that miR-125a-3p directly combines with circSND1, EGFP reporter assays were performed. As shown in Figure 3C, miR-125a-3p significantly decreased the EGFP intensity of circSND1-WT. Nevertheless, miR-125a-3p had no effect on the EGFP intensity of circSND1-mut, which was mutated in the miR-125a-3p binding sites of circSND1. RT-qPCR results showed that overexpression of circSND1 decreased the level of endogenous miR-125a-3p while knockdown of circSND1 increased the level of miR-125a-3p (Figure 3D).
Functional experiments illustrated that overexpression of miR-125a-3p significantly inhibited migration and invasion ability, while ASO-miR-125a-3p enhanced the migration and invasion ability of CC cells (Figure 3E and F). Next, we checked the expression of key molecular markers in EMT (E-cadherin and vimentin) to explore the role of miR-125a-3p in this process. Western blot assay showed that miR-125a-3p increased the protein levels of E-cadherin but decreased vimentin (Figure 3G). Therefore, our results demonstrated that circSND1 served as miR-125-3p sponge and miR-125a-3p suppressed the migration and invasion ability and EMT process of HeLa and SiHa cells.
FUT6 is a Direct Target of miR-125a-3p and Enhances the Migration and Invasion in Cervical Cancer Cells
Previous research showed that circRNAs might be involved in tumor progression through circRNA–miRNA–mRNA pathway.27 To identify possible candidates that mediate the effects of miR-125a-3p in CC cells, we predicted with Targetscan (http://www.targetscan.org) and selected a target protein, FUT6, which contained conserved target sites of miR-125a-3p (Figure 4A). We constructed the EGFP reporter plasmids containing 3′ UTR of FUT6. As indicated in Figure 4B, miR-125a-3p over-expression weakened the EGFP expression, while miR-125a-3p inhibition enhanced the expression of EGFP in the wild type reporter. However, these effects disappeared once the binding sites of miR-125a-3p was mutated. RT-qPCR and Western blot assays showed that miR-125a-3p significantly decreased the endogenous FUT6 expression while miR-125a-3p silencing elevated the expression of FUT6 (Figure 4C). In HeLa and SiHa cells, overexpression of FUT6 promoted, but knockdown of FUT6 inhibited cell migration and invasion ability (Figure 4D). Meanwhile overexpression of FUT6 promoted the EMT progress with increasing vimentin and decreasing E-cadherin (Figure 4E). In order to further confirm that miR-125-3p inhibits the malignant behavior of CC cells through FUT6, we conducted a rescue experiment. After co-transfection of FUT6 with miR-125a-3p, the inhibitory effect of miR-125a-3p on migration and invasion was weakened in HeLa and SiHa cells compared to the miR-125a-3p transfected group (Figure 4F). Western blot analysis showed that the inhibition of miR-125a-3p on EMT process was reduced compared to the miR-125a-3p transfected group (Figure 4G). FUT6 was indicated by these data as a direct downstream target of miR-125a-3p in cervical cancer cells, which facilitated migration, invasion and EMT process in CC cells.
CircSND1 Promotes the Progression of CC via the circSND1/miR-125a-3p/FUT6/NF-κB Axis
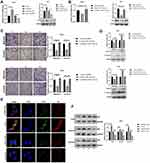
To determine whether circSND1 regulates CC progression through miR-125a-3p/FUT6 axis, we detected the effect of circSND1 on FUT6 expression first and found that circSND1 over-expression upregulated FUT6 expression on mRNA and protein levels while circSND1 knockdown downregulated it (Figure 5A). Subsequently, we performed rescue assays. The inhibitory effect of miR-125a-3p on FUT6 expression was counteracted by circSND1 overexpression as manifested in RT-qPCR and Western blot assays (Figure 5B). And similar results were observed in functional rescue experiments. As shown in (Figure 5C), circSND1 can restore the inhibitory effect of miR-125a-3p on migration and invasion in HeLa and SiHa cells. The Western blot of EMT markers showed similar result (Figure 5D). Our data demonstrated that circSND1 is endogenously competitive for miR-125a-3p to increase the expression of FUT6, thus facilitating migration/invasion and EMT process in CC cells.
Our results revealed that TNF-α promoted the expression of circSND1 via NF-κB and then circSND1 promoted the expression of FUT6 through the ceRNA mechanism. In recent research, FUT6 significantly enhanced the activation of PI3K/Akt signaling pathway and promoted the expression of p65.28,29 Therefore, we examined whether FUT6 had an effect on NF-κB activation in HeLa cells. Immunofluorescence analysis showed that the nucleus accumulation of p65 was obviously increased after FUT6 transfection, but inhibited by pshR-FUT6 transfection (Figure 5E). Simultaneous, nuclear cytoplasmic protein separation assay demonstrated that overexpression of FUT6 increased the level of p65 in nucleus whereas knockdown of p65 decrease its level (Figure 5F). Therefore, circSND1 exerted its functions via the TNF-α/NF-κB/circSND1/miR-125a-3p/FUT6/NF-κB positive regulatory circuit and promoted the malignant behavior of CC cells.
CircSND1 Accelerates Metastasis of Xenograft Tumors in vivo
We constructed the human CC xenograft model in the nude mice to study the influence of circSND1 on tumor metastasis in vivo. HeLa cells with stable overexpression of circSND1 and empty vector were injected subcutaneously into the tail vein of female nude mice. The results showed that the overexpression of circSND1 promoted CC malignant behaviors obviously with more lung metastatic nodules and shorter survival time (Figure 6A–C). IHC result showed that the expression of FUT6 was increased remarkably in circSND1 overexpressing xenograft tumor tissues (Figure 6D). RT-qPCR and Western blot assay revealed that FUT6 was strongly increased after circSND1 overexpression (Figure 6E and F). This is consistent with the results from in vitro assays, showing that circSND1 plays the role of oncogene in CC.
In summary, our study showed that circSND1 promotes migration/invasion and EMT process of CC cells through TNF-α/NF-κB/circSND1/miR-125a-3p/FUT6/NF-κB axis (Figure 6G). The results from this study may present a new biomarker for the early diagnosis and treatment of cervical cancer.
Discussion
Cervical cancer is the fourth most common gynecological malignant tumor in the world. Almost all cervical cancers are caused by high-risk subtype HPV viruses, especially HPV-16 and HPV-18.30 Studies showed that the increase in the number of cervical epithelial macrophages will exacerbate HPV infection, leading to inflammatory reactions.31 After the early HPV infection, the HPV virus integrates certain parts of its genome into the genome of the host cells, thus causing the massive release of pro-inflammatory factors and inducing chronic inflammation.32 Therefore, continuous HPV infection and the chronic inflammation induced by it are the main reasons for the occurrence and development of cervical cancer.
In human transcripts, only 2% are encoded as proteins and most of the transcripts are termed as non-coding RNAs. As a major discovery in bioscience, the aberrant expression and role of non-coding RNAs in tumorigenesis and progression of tumors was extensively studied in recent years. Research results have demonstrated that lncRNA and microRNA play important roles in the process of inflammation-related cervical cancer development.33,34 However, only limited studies indicated the involvement of circRNAs in inflammation-related diseases. In our research, we firstly identified circSND1 as a novel and up-regulated circRNA upon TNF-α stimulation in HeLa cells. CircSND1 originated from the back-splice of SND1 exons 8, 9 and 10. And sanger sequencing, RNase R, PCR and agarose gel electrophoresis assays confirmed that circSND1 is indeed a circRNA. Next, to examine the regulation mechanism of circSND1 expression, we cloned a 1000 bp sequence corresponding to the human SND1 gene promoter into the firefly luciferase reporter vector pGL3-Basic. It contains a highly conserved 300 bp segment with NF-κB sites and other cis-regulatory elements.22 Proinflammatory cytokine TNF-α caused the activation of NF-κB cascade. NF-κB is the key transcription factor that induces the expression of protective genes and finally resolves the protection or death of the cell.35 To assess whether TNF-α promoted the expression of circSND1 via NF-κB, three NF-κB binding sites (at −116, −174 and −267) in a converged 300 bp region upstream to the transcription start site of the promoter sequences of SND1 were mutated and assayed for luciferase activity. Results showed that mutations for NF-κB binding at −116 (mut#1) or −174 (mut#2) appeared a decrease of SND1 promoter activity. No effect observed when NF-κB −322 (mut#3) was changed in the region of SND1 promoter may suggest that this is not important role for promoter function. In addition, TNF-α promoted the activity of wild type SND1 promoter and partially promoted the activities of mut#1 and mut#2 promoter; while TNF-α had no obvious effect on the activities of mut#3 promoter. Subsequently, the two NF-κB binding sites were simultaneously mutated (at −116 and −174), the promotion of TNF-α was abolished. Previous study demonstrated that there is a putative enhancer role for the cis-acting elements in the region from −274 to −112 of SND1 promoter, among them potential NF-κB binding sites (at −116 and −174) would be important for transcriptional activation control.22 Then, we assessed the induction of the p65 subunit of NF-κB on circSND1 promoter activity and found similar results with overexpressing of p65. RT-qPCR assay demonstrated that overexpression of p65 increased the expression of circSND1 and mSND1. And to further confirm TNF-α promotes the expression of circSND1 through NF-κB, we studied the effect of TNF-α on the expression of circSND1 in the pretreatment of the NF-κB inhibitor BAY 11–7082. Our results showed that blockage of canonical NF-κB signaling reduced the TNF-α induced circSND1 expression. These results showed that the up-expression of circSND1 was induced by TNF-α through activation of the NF-κB signal pathway in HeLa cells.
Next, functional analysis showed that circSND1 did not influence cell viability or colony formation ability in CC cells, but it promoted migration/invasion ability and EMT process. Additionally, in vivo experiments confirmed that circSND1 promoted the lung metastasis of CC. The results showed that circSND1 regulated by TNF-α has a key role in the progression of CC. HPVs are small, nonenveloped double-stranded DNA viruses. Oncoproteins encoded by E6 and E7 genes are important factors that cause cervical epithelial cancer. Both E6 and E7 contribute to achieve uncontrolled proliferation through deregulation of growth suppressors.23 To determine whether circSND1 has an effect on the two oncoproteins of HPV in the process of CC, we detected the expression levels of E6/E7 oncogenes after overexpression of circSND1 in HeLa and SiHa cells, with the fact that HeLa cells harboring HPV 18 type, SiHa harboring HPV16 type. Results showed that circSND1 had no obvious effect on the expression of E6 and E7.
CircRNA functions through the classic way of the ceRNA mechanism. Our study showed that miR-125a-3p inhibited the migration, invasion, and EMT processes, suggesting that miR-125a-3p played a tumor suppressor function in CC cells. We confirmed that circSND1 bound to miR-125a-3p and inhibited the expression of miR-125a-3p. Furthermore, FUT6 is a latent target of miR-125a-3p by using TargetScan (http://www.targetscan.org) for bioinformatics analysis. Subsequently, the Western blotting discovered that miR-125a-3p bound to the 3′ UTR of FUT6 directly in EGFP report assay and reduced the mRNA and protein levels of FUT6. Rescue assays further confirmed that FUT6 was the functional target gene of miR-125a-3p and miR-125a-3p inhibited the malignant behavior of CC cells through FUT6. The rescue experiments showed that circSND1 reduced the inhibitory effect of miR-125a-3p on FUT6. Therefore, circSND1 may act as a sponge for miR-125a-3p to regulate the expression of FUT6. FUT family is abnormally expressed in breast cancer36 and hepatocellular carcinoma.37 Gain/loss-of-function analysis revealed that altered FUT6 promoted migration/invasion and EMT process in vitro. Meanwhile, immunofluorescence analysis and nuclear cytoplasmic separation assay showed that over-expression of FUT6 accelerated the nuclear translocation of p65 and could activate the NF-κB signaling pathway.
In addition to the ceRNA mechanism, circRNA may also exert its effect through other mechanisms including protein encoding. CircRNA needs two basic elements for translation ability: open reading frame (ORF) and translation initiation element (IRES). Containing ORF regions but not IREs, circSND1 may not encode peptide. In the latest study, m6A methylation modification can also lead to circRNA-encoded peptides.38 We predicted by SRAMP (http://www.cuilab.cn/sramp) and found that there are m6A methylation modification sites in the circSND1 sequence. Hence, further study is needed to evaluate whether circSND1 can encode a protein.
In conclusion, our results demonstrated that circSND1 exerted its functions via the TNF-α/NF-κB/circSND1/miR-125a-3p/FUT6/NF-κB positive regulatory circuit and promoted the process of CC, which suggests that circSND1 can be a promising prognostic marker and therapeutical target for cervical cancer.
Abbreviations
CC, cervical cancer; CircRNA, circular RNA; TNF-α, tumor necrosis factor-α; NF-κB, nuclear factor-κB; EMT, epithelial‐mesenchymal transition; HPV, human papillomavirus; ceRNA, competing endogenous RNA.
Ethics Declarations
All animal experiment procedures are approved by the Animal Care and Use Committee of Tianjin Medical University and comply with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Funding
This work was supported by the National Natural Science Foundation of China (Numbers: 81830094, 91629302, 81572790, 81773002) and the Natural Science Foundation of Tianjin (16JCYBJC42400).
Disclosure
The authors declare no conflict of interest.
References
1. Castellsagué X. Natural history and epidemiology of HPV infection and cervical cancer. Gynecol Oncol. 2008;110(3 Suppl 2):S4–S7.
2. Crosbie EJ, Einstein MH, Franceschi S, et al. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889–899. doi:10.1016/S0140-6736(13)60022-7
3. Shi J-F, Canfell K, Lew J-B, et al. The burden of cervical cancer in China: synthesis of the evidence. Int J Cancer. 2012;130(3):641–652. doi:10.1002/ijc.26042
4. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
5. Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
6. Hsu M-T, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi:10.1038/280339a0
7. He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi:10.1016/j.canlet.2017.03.027
8. Li X, Yang L, Chen -L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
9. Li X-N, Wang Z-J, Ye C-X, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. doi:10.1186/s13046-018-1006-x
10. Taulli R, Loretelli C, Pandolfi PP. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol. 2013;20(5):541–543. doi:10.1038/nsmb.2580
11. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi:10.1016/j.jhep.2018.01.012
12. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1):27. doi:10.1186/s12943-019-0951-0
13. Elinav E, Nowarski R, Thaiss CA, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi:10.1038/nrc3611
14. Guan X, Zong Z-H, Liu Y, et al. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892. doi:10.1016/j.omtn.2019.09.032
15. Chen T, Yu Q, Xin L, et al. Circular RNA circC3P1 restrains kidney cancer cell activity by regulating miR-21/PTEN axis and inactivating PI3K/AKT and NF- k B pathways. J Cell Physiol. 2020;235(4):4001–4010. doi:10.1002/jcp.29296
16. Liu D, Conn V, Goodall GJ, et al. A highly efficient strategy for overexpressing circRNAs. Methods Mol Biol. 2018;1724:97–105.
17. Zhao JL, Zhang L, Guo X, et al. miR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life. 2015;67(5):380–394. doi:10.1002/iub.1381
18. Ren ZJ, Nong XY, Lv YR, et al. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis. 2014;5(8):e1387. doi:10.1038/cddis.2014.327
19. Chen Z, de Paiva CS, Luo LH, et al. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22(3):355–366.20. doi:10.1634/stemcells.22-3-355
20. Fan J-Y, Fan Y-J, Wang X-L, et al. miR-429 is involved in regulation of NF-κBactivity by targeting IKKβ and suppresses oncogenic activity in cervical cancer cells. FEBS Lett. 2017;591(1):118–128. doi:10.1002/1873-3468.12502
21. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1(6):a001651. doi:10.1101/cshperspect.a001651
22. Armengol S, Arretxe E, Rodríguez L, et al. NF-κB, Sp1 and NF-Y as transcriptional regulators of human SND1 gene. Biochimie. 2013;95(4):735–742. doi:10.1016/j.biochi.2012.10.029
23. Hoppe-Seyler K, Bossler F, Braun JA, et al. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26(2):158–168. doi:10.1016/j.tim.2017.07.007
24. Pal A, Kundu R. Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front Microbiol. 2020;10:3116. doi:10.3389/fmicb.2019.03116
25. Gu Z, Long J, Li Y, et al. MiR-125a-3p negatively regulates osteoblastic differentiation of human adipose derived mesenchymal stem cells by targeting Smad4 and Jak1. Am J Transl Res. 2019;11(4):2603–2615.
26. Zhang Y, Chen X, Deng Y. miR-125a-3p decreases levels of interleukin-17 and suppresses renal fibrosis via down-regulating TGF-β1 in systemic lupus erythematosus mediated lupus nephritic mice. Am J Transl Res. 2019;11(3):1843–1853.
27. Miao L, Yin RX, Zhang QH, et al. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Sci Rep. 2019;9(1):18314. doi:10.1038/s41598-019-54603-2
28. Liang L, Gao C, Li Y, et al. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8(8):e2968. doi:10.1038/cddis.2017.352
29. Cheng L, Luo S, Jin C, et al. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 2013;4(11):e923. doi:10.1038/cddis.2013.450
30. Biswas A. Human papillomavirus (HPV) and cervical cancer. J Indian Med Assoc. 2000;98(2):53–55.
31. Hammes LS, Tekmal RR, Naud P, et al. Macrophages, inflammation and risk of cervical intraepithelial neoplasia (CIN) progression–clinicopathological correlation. Gynecol Oncol. 2007;105(1):157–165. doi:10.1016/j.ygyno.2006.11.023
32. Liu X, Ma X, Lei Z, et al. Chronic inflammation-related HPV: a driving force speeds oropharyngeal carcinogenesis. PLoS One. 2015;10(7):e0133681. doi:10.1371/journal.pone.0133681
33. Fan Y, Nan Y, Huang J, et al. Up-regulation of inflammation-related LncRNA-IL7R predicts poor clinical outcome in patients with cervical cancer. Biosci Rep. 2018;38(3):BSR20180483. doi:10.1042/BSR20180483
34. Bumrungthai S, Ekalaksananan T, Evans MF, et al. Up-regulation of miR-21 is associated with cervicitis and human papillomavirus infection in cervical tissues. PLoS One. 2015;10(5):e0127109. doi:10.1371/journal.pone.0127109
35. Pikarsky E, Porat RM, Stein I, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–466. doi:10.1038/nature02924
36. Li N, Liu Y, Miao Y, et al. MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB Life. 2016;68(9):764–775. doi:10.1002/iub.1541
37. Cheng L, Gao S, Song X, et al. Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs. Oncotarget. 2016;7(38):61199–61214. doi:10.18632/oncotarget.11284
38. Zhang L, Hou C, Chen C, et al. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19(1):105. doi:10.1186/s12943-020-01224-3
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.