Back to Journals » Cancer Management and Research » Volume 12
CircSEC31A Promotes the Malignant Progression of Non-Small Cell Lung Cancer Through Regulating SEC31A Expression via Sponging miR-376a
Authors Cheng F, Yu J, Zhang X, Dai Z, Fang A
Received 2 September 2020
Accepted for publication 3 October 2020
Published 10 November 2020 Volume 2020:12 Pages 11527—11539
DOI https://doi.org/10.2147/CMAR.S280124
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Fengfeng Cheng, Jing Yu, Xiaoying Zhang, Zongyan Dai, Aiju Fang
Department of Pathology, Shandong Provincial Third Hospital, Cheeloo College of Medicine, Shandong University, Jinan, Shandong 250031, People’s Republic of China
Correspondence: Aiju Fang
Department of Pathology, Shandong Provincial Third Hospital, Cheeloo College of Medicine, Shandong University, 11 Wuyingshan Zhong Road, Tianqiao District, Jinan, Shandong 250031, People’s Republic of China
Tel +86-15550024278
Email [email protected]
Background: Circular RNAs (circRNAs) have recently been shown as important regulators in the pathogenesis of non-small cell lung cancer (NSCLC). The purpose of this work was to explore the precise parts played by circRNA SEC31 homolog A (circSEC31A, hsa_circ_0001421) in NSCLC malignant progression.
Methods: The expression levels of circSEC31A, miR-376a and SEC31 homolog A (SEC31A) were gauged by quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot. Subcellular fractionation assay was used to determine the subcellular localization of circSEC31A, and RNase R assay was performed to assess the stability of circSEC31A. Cell migration and invasion were detected by transwell assay, and cell apoptosis was evaluated using flow cytometry. Measurement of glucose consumption, lactate production and adenosine triphosphate (ATP) level were done using corresponding assay kits. The targeted interactions among circSEC31A, miR-376a and SEC31A were confirmed by the dual-luciferase reporter and RNA pull-down assays. Animal studies were performed to observe the role of circSEC31A in tumor growth in vivo.
Results: Our data indicated that circSEC31A and SEC31A were upregulated in NSCLC tissues and cells. CircSEC31A knockdown suppressed NSCLC cell migration, invasion, glycolysis and promoted apoptosis in vitro, as well as hindered tumor growth in vivo. Mechanistically, circSEC31A directly interacted with miR-376a, and circSEC31A depletion regulated NSCLC cell malignant progression by miR-376a. Moreover, SEC31A was a functional target of miR-376a, and it mediated the regulatory impact of miR-376a overexpression on NSCLC cell progression. Furthermore, circSEC31A controlled SEC31A expression through acting as a miR-376a sponge.
Conclusion: Our findings first identified that the knockdown of circSEC31A suppressed NSCLC malignant progression at least partly through modulating SEC31A expression by acting as a miR-376a sponge, providing a novel molecular target of NSCLC therapy.
Keywords: NSCLC, circSEC31A, miR-376a, SEC31A, malignant progression
Introduction
Lung cancer is the most prevalent malignancy and the most common cause of cancer death worldwide,1 of which non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer cases.2 Despite remarkable advances in NSCLC treatment, the long-term survival of these patients is still unsatisfactory.3 This highlights an urgent need to develop novel diagnostic biomarkers and effective therapies for NSCLC.
Being covalently closed, circular RNAs (circRNAs) are emerging participators in cancer biology.4 By serving as natural microRNA (miRNA) sponges, numerous circRNAs have been demonstrated as crucial regulators in NSCLC pathogenesis.5,6 For instance, Chen et al reported that circ_100146 contributed to NSCLC malignant progression by directly binding to miR-615-5p and miR-361-3p.7 Wang et al underscored that circ_0008305 protected against the inhibition of transcriptional intermediary factor 1γ to serve as a potential promoter in NSCLC metastasis via sponging miR-429/miR-200b-3p.8 Cui et al illuminated that circ_0043278 functioned as a miR-520f sponge to enhance NSCLC development.9
SEC31 homolog A (SEC31A), an outer component of coat protein complex II (COPII), has been illuminated to be linked to NSCLC pathogenesis.10,11 More importantly, circRNA SEC31A (circSEC31A, hsa_circ_0001421), derived from the backsplice of SEC31A gene, was found to be overexpressed in NSCLC tumor tissues and contribute to NSCLC progression.12 Although this report also uncovered a regulatory mechanism of circSEC31A in NSCLC, our understanding of its molecular basis is still limited. Intriguingly, when we utilized computer algorithms to identify the molecular basis underlying the involvement of circSEC31A in NSCLC, we found a potential regulatory network, the circSEC31A/miR-376a/SEC31A axis. Previous work had demonstrated the anti-tumor role of miR-376a (also called miR-376a-3p) in NSCLC tumorigenesis.13,14
Here, we investigated the involvement of circSEC31A in NSCLC malignant progression. We identified circSEC31A as a sponge of miR-376a to influence SEC31A expression, thereby regulating NSCLC cell malignant behaviors in vitro and in vivo.
Materials and Methods
Bioinformatics
The expression pattern of SEC31A was evaluated by the starbase v.3 software in 526 lung adenocarcinoma specimens and 59 normal controls. Analysis for the miRNAs that potentially bind to circSEC31A was performed using online softwares starbase v.3 and circbank. The molecular targets of miR-376a were predicted using the microT-CDS software.
Clinical Samples and Cells
The tumor samples and the matched healthy tissues of 44 patients with primary NSCLC, who had undergone pneumonectomy in Shandong Provincial Third Hospital, Shandong University from May 2015 to August 2017, were studied. The detailed information of these patients was provided in Table 1. The study protocol was approved by the Ethics Committee of Shandong Provincial Third Hospital, Shandong University, and informed consent was provided by all subjects before the start of the study.
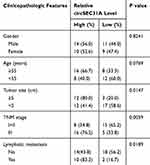 |
Table 1 Correlation of circSEC31A Expression with the Clinicopathological Characteristics of the NSCLC Patients |
Human normal 16HBE cells and A549 and H1299 NSCLC cells were obtained from Bnbio (Beijing, China) and propagated in RPMI-1640 medium (Gibco, Lofer, Austria) supplemented with 10% Gibco fetal bovine serum (FBS) under standard conditions.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA isolated from tissues and cells with Tri-reagent (Invitrogen, Toyko, Japan) averaged 650 ng/mL when quantified using a spectrophotometer (HITACHI, Beijing, China). For circSEC31A and mRNA levels, RNA (10 ng) was reverse-transcribed into cDNA using QuantiTect RT Kit (Qiagen, Hombrechtikon, Switzerland) and then analyzed using qRT-PCR (Qiagen SYBR Green) and specific primers (provided in Table 1) in triplicate. For miRNA expression, cDNA was synthesized using Qiagen miScript RT Kit and then subjected to qRT-PCR with miScript SYBR Green and primers for miRNA (provided in Table 2) in triplicate. Fold changes were determined by the comparative Ct (2−ΔΔCt) method, normalizing the results to the expression of β-actin or U6.
 |
Table 2 Sequences Used for PCR Primers and Oligonucleotides |
Western Blot
Protein extracts were prepared using HEPES lysis buffer and quantified with the BCA assay (Thermo Fisher Scientific, Runcorn, UK). After immunoblotting, the proteins were transferred onto a Hybond polyvinylidene fluoride membrane (GE Healthcare, Little Chalfont, UK). Following the probe with anti-SEC31A (1:2000, Invitrogen) or anti-β-actin (1:3000, Invitrogen) antibody, the immunocomplexes were treated with secondary antibody (1:10,000, Invitrogen) before being visualized by the enhanced Chemiluminescence (GE Healthcare).
Subcellular Fractionation Assay
RNA was obtained from the cytoplasm and nucleus of H1299 and A549 cells, respectively, using Cytoplasmic & Nuclear RNA Purification Kit based on the guidance of manufacturers (Norgen Biotek, Thorold, ON, Canada). Each measurement was done with 1 ng of RNA, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls.
RNase R Assay
This assay was carried out by adding 10 U of RNase R (GENESEED, Guangzhou, China) into 2.5 µg of RNA and 20 min incubation at 37°C.
Transient Transfection of Cells
CircSEC31A overexpression plasmid was created by cloning its full-length sequence into the pCD-ciR vector (GENESEED) with EcoR I and BamH I sites. SEC31A overexpression plasmid was generated by inserting human SEC31A sequence into the pcDNA3.1 vector (Invitrogen) opened with Not I and Xba I. Subconfluent cells in 24-well plates were transiently transfected with 60 nM of siRNAs targeting circSEC31A (si-circSEC31A#1, si-circSEC31A#2 or si-circSEC31A#3) or a nontarget siRNA control (si-con), 200 ng of the indicated plasmids, 30 nM of miR-376a mimic or negative mimic control (miR-con mimic), the inhibitor of mature miR-376a (in-miR-376a) or corresponding oligonucleotide control (in-miR-con) using the Lipofectamine 3000 (Invitrogen) following the suggestion of manufacturers. All oligonucleotides were obtained from GenePharma (Shanghai, China) and their sequences were shown in Table 2.
Transwell Migration and Invasion Assays
24-Transwell chambers (Corning, Tewksbury, MA, USA) with non-coated insert membranes (8-µm pore size) and Matrigel-coated membrane inserts were used for transwell migration and invasion assays, respectively. Transfected cells in 0.5% FBS medium were seeded into the top chamber, and media containing 15% FBS was used as a chemoattractant in the bottom. 24 h later, the cells that had migrated or invaded to the lower surface of the membrane were counted under a microscope at 100× magnification after being stained with 1% crystal violet.
Flow Cytometry
After 48 h transfection, about 10,000 cells were resuspended in a 500 µL volume of binding buffer containing 1.25 µL of Annexin V-FITC and 10 µL of propidium iodide based on the directions of the Apoptosis Kit (Elabscience, Wuhan, China). A BD flow cytometer (BD Biosciences, Heidelberg, Germany) was employed for data analysis.
Determination of Glucose Consumption, Lactate Production and Adenosine Triphosphate (ATP) Level
These were performed using the Colorimetric glucose consumption, lactate production, and ATP level Assay Kits, respectively, as per the recommendation of manufacturers (Elabscience).
RNA Pull-Down Assay
Biotin-labeled circSEC31A and the segment of SEC31A 3ʹUTR harboring miR-376a-matched region (circSEC31A probe and SEC31A probe) were obtained from VIAGENE (Shanghai, China), with the nontarget sequence labeled by biotin as the negative control. Cells were homogenized in lysis buffer and then incubated with circSEC31A probe or SEC31A probe for 3 h at 4°C before adding Streptavidin agarose beads (GlpBio, Shanghai, China) for 3 h. Subsequently, total RNA was extracted from the beads and then subjected to qRT-PCR for gene expression.
Dual-Luciferase Reporter Assay
The segments of circSEC31A and SEC31A 3ʹUTR harboring miR-376a-matched region or miss-matched target sites were individually inserted into pmirGLO plasmid (Promega, Southampton, UK). 50 ng of the indicated luciferase reporter was introduced into NSCLC cells with the mimic of miR-con or miR-376a at a final concentration of 30 nM. Luciferase activities were determined using the Promega Dual-Luciferase Assay Kit after 24 h transfection.
Generation of Stable Cell Line
shRNA lentiviruses encoding sequence targeting circSEC31A (sh-circSEC31A) or nontarget control (sh-con) were purchased from HANBIO (Shanghai, China). For lentiviral transduction, A549 cells of 50% confluence were infected by virus particles in media containing 8 µg/mL of polybrene. 24 h later, the vector-positive cells were selected by 1 µg/mL of puromycin over 72 h.
Animal Studies
All protocols for animal studies were approved by the Animal Care and Use Ethics Committee of Shandong Provincial Third Hospital, Shandong University, and all animal procedures were performed according to the Academia Sinica IACUC and Council of Agriculture Guidebook for the Care and Use of Laboratory Animals. About 1 × 107 A549 cells transduced with or without sh-circSEC31A or sh-con were implanted into the right side of each male BALB/c nude mice (Shanghai Animal Laboratory Center, Shanghai, China; n = 6 each group) by subcutaneous injection. The volume of the tumors (length × width2 × 0.5) was monitored every week and the tumors were excised at the end of the experiment.
Statistical Analysis
Unless otherwise indicated, all data were analyzed using a Student’s t-test, Mann–Whitney U-test or analysis of variance (ANOVA). Data were presented as the mean ± standard deviation. The correlations among circSEC31A, miR-376a and SEC31A in tumor samples were evaluated by the Spearman correlation test. The P values were *P < 0.05, **P < 0.01 or ***P < 0.001.
Results
CircSEC31A and SEC31A Were Overexpressed in NSCLC Tissues and Cells
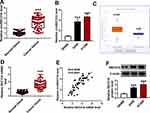
For preliminary investigation for the involvement of circSEC31A in NSCLC pathogenesis, we firstly determined its expression pattern in NSCLC tissues and cells. As demonstrated by qRT-PCR, in contrast to their counterparts, circSEC31A was significantly overexpressed in NSCLC tissues and cells (Figure 1A and B). Using the starBase v.3 software, a significant up-regulation of SEC31A was discovered in lung cancer tissues compared to the normal controls (Figure 1C). We then assessed the level of SEC31A in NSCLC tissues and cells. The data revealed that SEC31A expression was higher in NSCLC tissues than that of control (Figure 1D). Of interest, a positive correlation between SEC31A level and circSEC31A expression was found in NSCLC tissues (Figure 1E). Likewise, SEC31A expression was remarkably elevated in NSCLC cells compared with 16HBE cells (Figure 1F). Additionally, as shown in Table 1, circSEC31A expression was closely associated with tumor size, TNM stage and lymphatic metastasis of these patients.
Knockdown of circSEC31A Restrained NSCLC Cell Migration, Invasion, Glycolysis and Enhanced Cell Apoptosis in vitro
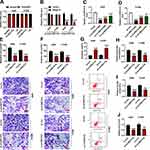
To determine the subcellular localization of circSEC31A in NSCLC cells, we performed subcellular fractionation assays. These results showed that circSEC31A was mainly localized in the cytoplasm of A549 and H1299 cells (Figure 2A). Moreover, the incubation of RNase R led to a prominent reduction in the level of SEC31A linear mRNA, and circSEC31A was resistant to RNase R (Figure 2B), demonstrating the inherent stability of circSEC31A.
To elucidate the biological effect of circSEC31A on NSCLC progression, we then carried out in vitro loss-of-function analyses by silencing circSEC31A using specific siRNAs (si-circSEC31A#1, si-circSEC31A#2 or si-circSEC31A#3). Transient transfection of siRNAs, but not the negative si-con control, significantly down-regulated the expression of circSEC31A in the two NSCLC cell lines (Figure 2C and D). si-circSEC31A#1 caused the most significant reduction in circSEC31A level, so we selected it (also called si-circSEC31A) for further analyses. Transwell assays showed that circSEC31A depletion induced a striking repression in cell migration (Figure 2E) and invasion (Figure 2F). Conversely, the silencing of circSEC31A in both the cell lines caused a prominent promotion in cell apoptosis compared to the negative control (Figure 2G). Furthermore, circSEC31A knockdown resulted in decreased levels of glucose consumption (Figure 2H), lactate production (Figure 2I) and ATP (Figure 2J) in the two NSCLC cell lines, indicating the suppression of circSEC31A knockdown on cell glycolysis.
CircSEC31A Directly Interacted with miR-376a via Binding to miR-376a
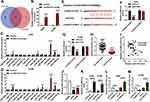
To further understand the role of circSEC31A in NSCLC, we performed a detailed analysis for the miRNAs that potentially bind to circSEC31A. Using both starbase and circbank softwares, the predicted data showed that circSEC31A harbored a region that matches the target sequences of 13 miRNAs (Figure 3A). To identify the direct interaction between circSEC31A and the 13 candidate miRNAs, we subsequently performed RNA pull-down assays with circSEC31A-specific probe. In comparison to the negative control, circSEC31A enrichment level was significantly elevated by circSEC31A probe in the two NSCLC cell lines (Figure 3B). Moreover, circSEC31A probe led to a remarkable increase in the enrichment levels of miR-376b, miR-383-5p, miR-376a and miR-525-5p in both two cell lines (Figure 3C and D). Among the four candidates, miR-376a was the most significantly elevated miRNA, so we assumed it as a targeted miRNA of circSEC31A. To address this possibility, we cloned the circSEC31A fragment into a luciferase plasmid and mutated the miR-376a-matched sites (Figure 3E). When we performed dual-luciferase reporter assays, the cotransfection of wild-type reporter construct (circSEC31A-WT) and miR-376a mimic into the two cell lines produced lower luciferase activity than cells cotransfected with miR-con control (Figure 3F and G). However, when the target sites were mutated (circSEC31A-MUT), little reduction was observed in luciferase activity in the presence of miR-376a mimic (Figure 3F and G). In NSCLC tissues, miR-376a expression was prominently reduced and inversely correlated with circSEC31A level (Figure 3H and I). Additionally, in contrast to the normal control, miR-376a was prominently underexpressed in both A549 and H1299 cells (Figure 3J). To elucidate whether circSEC31A regulated miR-376a expression, we manipulated circSEC31A level in the two NSCLC cells. The transfection efficiency of circSEC31A overexpressing plasmid was gauged by qRT-PCR (Figure 3K). As expected, miR-376a expression was strikingly decreased by circSEC31A overexpressing plasmid, while it was remarkably increased following circESC31A silencing in both the cells (Figure 3L and M).
CircSEC31A Regulated NSCLC Cell Migration, Invasion, Glycolysis and Apoptosis in vitro by miR-376a
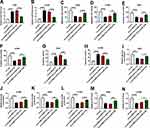
We then examined whether miR-376a was a molecular mediator of circSEC31A in NSCLC progression. As shown by qRT-PCR, the transfection of in-miR-376a notably abated si-circSEC31A-caused miR-376a upregulation in the two NSCLC cell lines (Figure 4A and B). Functional analyses data revealed that the reduced level of miR-376a strikingly abolished circSEC31A knockdown-induced anti-migration (Figure 4C and D), anti-invasion (Figure 4E and F) and pro-apoptosis (Figure 4G and H) effects. Furthermore, miR-376a downregulation remarkably abrogated the reduced effects of circSEC31A depletion on glucose consumption (Figure 4I and J), lactate production (Figure 4K and L) and ATP levels (Figure 4M and N) in both cell lines.
SEC31A in NSCLC Cells Was Directly Targeted and Inhibited by miR-376a
Using the microT-CDS software, a potential target sequence for miR-376a was discovered within the 3ʹUTR of SEC31A (Figure 5A). To validate this, we used SEC31A 3ʹUTR reporter constructs (SEC31A-WT or SEC31A-MUT) containing wild-type or mutant-type miR-376a-matched sites in dual-luciferase assays. Transient introduction of SEC31A-WT in the presence of miR-376a mimic caused a significant reduction in luciferase activity, and this effect was dramatically abrogated by SEC31A-MUT (Figure 5B and C). RNA pull-down data revealed that in contrast to the control group, the enrichment levels of SEC31A mRNA and miR-376a were synchronously elevated by SEC31A probe in the two NSCLC cell lines (Figure 5D–F). Of interest, in NSCLC tissues, a strong inverse correlation between SEC31A mRNA expression and miR-376a level was found (Figure 5G). Furthermore, the data of Western blot showed that SEC31A expression was strikingly decreased by miR-376a overexpression and increased as a result of miR-376a depletion in the two cell lines (Figure 5H and I).
MiR-376a Overexpression Hampered NSCLC Cell Migration, Invasion, Glycolysis and Promoted Apoptosis in vitro by Down-Regulating SEC31A
In addition to the reduced impact on SEC31A expression (Figure 6A and B), the elevated expression of miR-376a led to a distinct inhibition in cell migration (Figure 6C and D), invasion (Figure 6E and F), and a clear enhancement in cell apoptosis (Figure 6G and H), as well as a strong downregulation in glucose consumption (Figure 6I and J), lactate production (Figure 6K and L) and ATP level (Figure 6M and N). We then asked whether SEC31A was involved in miR-376a-mediated regulation on NSCLC malignant progression. As shown by Western blot, the transfection of SEC31A overexpressing plasmid dramatically abolished miR-376a overexpression-mediated SEC31A down-regulation (Figure 6A and B). Functional experiments results revealed that SEC31A expression restoration remarkably abrogated miR-376a overexpression-induced anti-migration (Figure 6C and D) and anti-invasion (Figure 6E and F), pro-apoptosis (Figure 6G and H) and anti-glycolysis (Figure 6I–N) effects.
CircSEC31A Controlled SEC31A Expression Through Sponging miR-376a
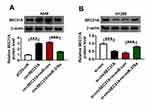
We next undertook to determine whether, if so, how circSEC31A regulated SEC31A in NSCLC cells. By contrast, in both cell lines, SEC31A protein expression was significantly elevated by circSEC31A overexpressing plasmid (Figure 7A), and it was prominently reduced when circSEC31A knockdown (Figure 7B). Nevertheless, the regulatory impact of circSEC31A on SEC31A expression was significantly abrogated by miR-376a level alteration in the two NSCLC cell lines (Figure 7A and B).
Knockdown of circSEC31A Weakened Tumor Growth in vivo
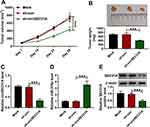
A crucial question was whether circSEC31A could regulate tumor growth in vivo. To address the possibility, we implanted sh-circSEC31A-transduced or sh-con-infected A549 cells into the nude mice to generate the xenograft model. In comparison to the negative control, the transduction of sh-circSEC31A caused a remarkable reduction in tumor growth (Figure 8A and B). Furthermore, the data of qRT-PCR and Western blot showed that circSEC31A and SEC31A were significantly underexpressed and miR-376a was strikingly overexpressed in the tumor tissues derived from the sh-circSEC31A-transduced A549 cells (Figure 8C–E).
Discussion
Accumulating evidence is pointing towards circRNAs as key players in NSCLC pathogenesis, highlighting a novel field of diagnostic and therapeutic opportunities.6,15 However, identifying the precise actions of these circRNAs in NSCLC pathogenesis has been challenging. In this work, we investigated the precise, critical parts of circSEC31A in NSCLC progression.
Here, we demonstrated the remarkable overexpression of circSEC31A in NSCLC, consistent with previous work.12 As has been reported for other circRNAs,16,17 circSEC31A was unusually stable and resistant to RNase R, owing to the lack of both 5ʹ and 3ʹ ends.18 As a hallmark of cancer biology, the enhanced glycolysis can satisfy the increasing energy requirement of cancer cells and promote cancer development.19 By a series of functional analyses, we identified the oncogenic activity of circSEC31A in NSCLC in vitro and in vivo. Our data also demonstrated the mainly cytoplasmic localization of circSEC31A in NSCLC cells, providing a possibility for its interaction with miRNAs.4
It is widely accepted that many circRNAs exert crucial functions in cancer biology by working as miRNA inhibitors.20 For the first time, we validated that circSEC31A in NSCLC cells directly interacted with miR-376a by binding to miR-376a. Work in a number of laboratories has indicated that as a member of 14q32 miRNA clusters, miR-376a can function as a strong tumor suppressor in many human cancers, such as giant cell tumor of bone, osteosarcoma and melanoma.21–23 On the contrary, miR-376a has been testified to be overexpressed and work as a potential promoter in ovarian cancer and papillary thyroid cancer.24–26 Moreover, miR-376a was reported to associate with poor outcome after resection in lung adenocarcinoma.27 Here, we validated the anti-tumor activity of the increased miR-376a expression in NSCLC, as has been reported.13 More importantly, we first identified miR-376a as a molecular mediator of circSEC31A on NSCLC malignant progression.
In NSCLC cells, the circRNA/miRNA/mRNA regulatory network has been most frequently reported.28–30 In this report, we were first to highlight that miR-376a in NSCLC cells directly targeted SEC31A, which was verified to be significantly overexpressed in NSCLC. SEC31A-anaplastic lymphoma kinase (ALK) fusion gene was implicated in NSCLC pathogenesis.10 Additionally, SEC31A mutation was discovered to have relevance to afatinib resistance in NSCLC.11 For the first time, our data uncovered that the elevated expression of miR-376a exerted a repressive effect in NSCLC progression by SEC31A. More interestingly, we first demonstrated that circSEC31A controlled SEC31A expression via sponging miR-376a. Animal studies showed that circSEC31A depletion resulted in increased miR-376a level and decreased SEC31A expression in the tumor tissues. Hence, more explorations about the new regulatory mechanism in vivo will be done in further work.
In conclusion, to our knowledge, this is the first report that the reduced circSEC31A level hampered NSCLC malignant progression via modulating SEC31A expression by working as a miR-376a sponge. A further challenge will be to identify how the novel network influences NSCLC cell malignant behaviors in vitro and in vivo.
Highlights
(1) CircSEC31A directly interacted with miR-376a through binding to miR-376a.
(2) CircSEC31A controlled SEC31A expression through acting as a miR-376a sponge.
(3) CircSEC31A knockdown suppressed NSCLC malignant progression through regulating the miR-376a/SEC31A axis.
Funding
The present study was supported by Shandong Province Key R&D Program Project (2018GSF1221040).
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454.
3. Sekihara K, Hishida T, Yoshida J, et al. Long-term survival outcome after postoperative recurrence of non-small-cell lung cancer: who is ‘cured’ from postoperative recurrence? Eur J Cardiothorac Surg. 2017;52(3):522–528. doi:10.1093/ejcts/ezx127
4. Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
5. Xu N, Chen S, Liu Y, et al. Profiles and bioinformatics analysis of differentially expressed circrnas in taxol-resistant non-small cell lung cancer cells. Cell Physiol Biochem. 2018;48(5):2046–2060. doi:10.1159/000492543
6. Van Der Steen N, Lyu Y, Hitzler AK, et al. The circular RNA landscape of non-small cell lung cancer cells. Cancers (Basel). 2020;12(5). doi:10.3390/cancers12051091.
7. Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):13. doi:10.1186/s12943-019-0943-0
8. Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. doi:10.1186/s12943-018-0889-7
9. Cui J, Li W, Liu G, et al. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47(1):810–821. doi:10.1080/21691401.2019.1575847
10. Kim RN, Choi YL, Lee MS, et al. SEC31A-ALK fusion gene in lung adenocarcinoma. Cancer Res Treat. 2016;48(1):398–402. doi:10.4143/crt.2014.254
11. Van Der Wekken AJ, Kuiper JL, Saber A, et al. Overall survival in EGFR mutated non-small-cell lung cancer patients treated with afatinib after EGFR TKI and resistant mechanisms upon disease progression. PLoS One. 2017;12(8):e0182885. doi:10.1371/journal.pone.0182885
12. Jin M, Shi C, Hua Q, et al. High circ-SEC31A expression predicts unfavorable prognoses in non-small cell lung cancer by regulating the miR-520a-5p/GOT-2 axis. Aging. 2020;12(11):10381–10397. doi:10.18632/aging.103264
13. Wang Y, Cong W, Wu G, et al. MiR-376a suppresses the proliferation and invasion of non-small-cell lung cancer by targeting c-Myc. Cell Biol Int. 2018;42(1):25–33. doi:10.1002/cbin.10828
14. Joerger M, Baty F, Früh M, et al. Circulating microRNA profiling in patients with advanced non-squamous NSCLC receiving bevacizumab/erlotinib followed by platinum-based chemotherapy at progression (SAKK 19/05). Lung Cancer. 2014;85(2):306–313. doi:10.1016/j.lungcan.2014.04.014
15. Wang C, Tan S, Liu WR, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134. doi:10.1186/s12943-019-1061-8
16. Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659.
17. Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36(11):901–908. doi:10.1089/dna.2017.3862
18. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
19. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi:10.1038/nrc3038
20. Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi:10.1016/j.trecan.2020.01.012
21. Herr I, Sähr H, Zhao Z, et al. MiR-127 and miR-376a act as tumor suppressors by in vivo targeting of COA1 and PDIA6 in giant cell tumor of bone. Cancer Lett. 2017;409:49–55. doi:10.1016/j.canlet.2017.08.029
22. Fellenberg J, Lehner B, Saehr H, et al. Tumor suppressor function of miR-127-3p and miR-376a-3p in osteosarcoma cells. Cancers (Basel). 2019;11(12). doi:10.3390/cancers11122019.
23. Zehavi L, Avraham R, Barzilai A, et al. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol Cancer. 2012;11(44). doi:10.1186/1476-4598-11-44.
24. Yang L, Wei QM, Zhang XW, et al. MiR-376a promotion of proliferation and metastases in ovarian cancer: potential role as a biomarker. Life Sci. 2017;173:62–67. doi:10.1016/j.lfs.2016.12.007
25. Meng X, Joosse SA, Müller V, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113(9):1358–1366. doi:10.1038/bjc.2015.340
26. Dai D, Tan Y, Guo L, et al. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur J Endocrinol. 2020;182(1):111–121. doi:10.1530/EJE-19-0524
27. Nadal E, Zhong J, Lin J, et al. A microRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res. 2014;20(12):3107–3117. doi:10.1158/1078-0432.CCR-13-3348
28. Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer. 2019;18(1):179. doi:10.1186/s12943-019-1111-2
29. Zhang H, Wang X, Hu B, et al. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol. 2019;47(1):3410–3416. doi:10.1080/21691401.2019.1652623
30. Li Y, Hu J, Li L, et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun. 2018;503(3):2089–2094. doi:10.1016/j.bbrc.2018.07.164
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.