Back to Journals » OncoTargets and Therapy » Volume 13
Circ_SATB2 Attenuates the Anti-Tumor Role of Celastrol in Non-Small-Cell Lung Carcinoma Through Targeting miR-33a-5p/E2F7 Axis
Authors Liu P, Wang M, Tang W, Li G, Gong N
Received 29 August 2020
Accepted for publication 21 October 2020
Published 18 November 2020 Volume 2020:13 Pages 11899—11912
DOI https://doi.org/10.2147/OTT.S279434
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Peijun Liu,* Miao Wang,* Weihua Tang, Guangcai Li, Nianjin Gong
Department of Respiratory and Critical Care Medicine, The Central Hospital of Enshi Tujia and Miao Autonomous Prefecture, Enshi, Hubei 445000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Nianjin Gong
Department of Respiratory and Critical Care Medicine, The Central Hospital of Enshi Tujia and Miao Autonomous Prefecture, No. 60, Hangkong Avenue, Enshi, Hubei 445000, People’s Republic of China
Tel +86-0718-8263451
Email [email protected]
Background: Celastrol (Cela) was a natural compound that exerted anti-tumor activity in many cancer cells. Nevertheless, the molecular mechanism behind the anti-tumor role of Cela in non-small-cell lung carcinoma (NSCLC) remains to be clarified.
Methods: Flow cytometry was used to analyze cell cycle progression and apoptosis. Colony formation assay and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay were used to analyze cell proliferation. Cell migration and invasion abilities were assessed by transwell assays. Quantitative real-time polymerase chain reaction (qRT-PCR) was implemented for the detection of RNA levels. Western blot assay was used for the determination of protein levels. Dual-luciferase reporter assay was conducted to confirm the interaction between microRNA-33a-5p (miR-33a-5p) and circular RNA SATB homeobox 2 (circ_SATB2) or E2F transcription factor 7 (E2F7). Xenograft tumor assay was conducted to test the roles of Cela and circ_SATB2 in NSCLC progression in vivo.
Results: Cela hampered the malignant behaviors of NSCLC cells. Cela down-regulated circ_SATB2 level in NSCLC cells. Cela stimulation-induced suppressive influence in NSCLC progression was alleviated by circ_SATB2 accumulation. E2F7 interference overturned circ_SATB2-mediated effects in Cela-stimulated NSCLC cells. MiR-33a-5p was a target of circ_SATB2, and E2F7 was verified as a target of miR-33a-5p. Circ_SATB2 attenuated Cela-mediated effects through targeting miR-33a-5p in NSCLC cells. Cela-mediated suppressive effect on tumor growth was partly attenuated by the overexpression of circ_SATB2 in vivo.
Conclusion: Cela suppressed NSCLC development through regulating circ_SATB2/miR-33a-5p/E2F7 signaling cascade.
Keywords: NSCLC, celastrol, circ_SATB2, miR-33a-5p, E2F7
Introduction
Lung cancer is a common malignancy that affects the health of both men and women globally.1,2 Non-small-cell lung carcinoma (NSCLC) occupies the majority of lung cancer cases.3,4 Due to the high metastasis and chemoresistance, the 5-year survival rate of NSCLC patients remains dismal.5,6 It is urgently needed to identify novel effective early diagnostic markers and therapeutic targets.
Celastrol (Cela) is a natural ingredient isolated from Tripterygium wilfordii, and it has been identified to exert an anti-tumor activity in diverse cancers, including NSCLC.7–9 Here, we identified a novel signal pathway by which Cela restrained the progression of NSCLC. Non-coding RNAs (ncRNAs) comprise about 98% of human genome.10 Circular RNAs (circRNAs), microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are both ncRNAs, and circRNAs are ideal biomarkers for human diseases because of their stable circular structure and crucial regulatory functions.11,12 CircRNAs function via diverse manners, including serving as miRNA sponges.13 For instance, circ_0001649 blocked NSCLC development by targeting miR-331-3p and miR-338-5p.14 As for circRNA SATB homeobox 2 (circ_SATB2), Zhang et al demonstrated that circ_SATB2 accelerated NSCLC development.15 However, the working mechanism of circ_SATB2 in NSCLC progression remains to be illustrated.
MiRNAs are small non-coding RNAs with about 22 nucleotides in length.16 MiRNAs have attached much attention because of their abilities to function as oncogenes or tumor suppressors in cancers. Wang et al found that circ_CCDC66 accelerated NSCLC development through targeting miR-33a-5p.17 However, the working mechanism of miR-33a-5p and the functional association between miR-33a-5p and Cela need to be clarified.
E2F transcription factor family was known to modulate multiple cellular behaviors.18 E2F transcription factor 7 (E2F7) was known as a transcription repressor. E2F7 was dysregulated in cancers, including cutaneous squamous cell carcinoma,19 ovarian cancer20 and NSCLC.21 Wang et al demonstrated that E2F7 was abnormally up-regulated in NSCLC tissues, and miR-935 hampered the proliferation, migration and invasion of NSCLC cells via targeting E2F7.21 Here, circ_SATB2/miR-33a-5p/E2F7 axis was firstly identified in NSCLC.
Cela suppressed the malignant behaviors of NSCLC cells, and circ_SATB2 expression was down-regulated with Cela stimulation in NSCLC cells. Subsequently, we further investigated if circ_SATB2 was involved in the suppressive role of Cela in NSCLC progression, and the downstream molecules of circ_SATB2 were explored.
Materials and Methods
Clinical Specimens
Forty-nine pairs of NSCLC tissues and adjacent non-tumor tissues were collected from 49 NSCLC patients at The Central Hospital of Enshi Tujia And Miao Autonomous Prefecture. All these subjects had not received preoperative chemotherapy, radiotherapy or other treatments. All of the subjects had signed written informed consents prior to the surgery, and the study protocol had gotten the permission of the Clinical Research Ethics Committee of The Central Hospital of Enshi Tujia And Miao Autonomous Prefecture.
Cell Culture
Five lung cancer cell lines (A549, H460, H1299, H226 and H522; BeNa Culture Collection, Beijing, China) and human bronchial epithelioid cell line 16HBE (BeNa Culture Collection), which were adherent cell lines, were cultured with Roswell Park Memorial Institute-1640 medium (RPMI-1640, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 10% penicillin/streptomycin at 37°C with 5% CO2 under saturation humidity.
Chemical and Treatment
Cela was obtained from Pie & Pie Technologies (Shenzhen, China). NSCLC cells treated with 1 μM or 3 μM Cela for 24 h were used for functional analysis.
Cell Cycle Analysis
Transfected NSCLC cells were collected using cold phosphate buffer saline (PBS; Sangon Biotech, Shanghai, China) and suspended in 70% ethanol solution at −20°C overnight. RNase was used to digest RNA content, and the DNA content was stained by propidium iodide (PI; Sigma, St. Louis, MO, USA). The cell cycle distribution was analyzed by flow cytometry.
Colony Formation Assay
NSCLC cells were seeded onto 6-well plates in triplicates at the density of 150 cells/well. Cells were cultivated for at least 2 weeks in the humidified incubator at 37°C with 5% CO2, and the culture medium was replenished every 4 d. Colonies were fixed using 4% poly methanol (Sangon Biotech) and then stained using crystal violet (Sangon Biotech). The images were pictured, and the number of colonies was counted with naked eyes.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
Logarithmic-phase NSCLC cells were transfected with oligonucleotides or plasmids. After transfection for 0 d, 1 d, 2 d or 3 d, 10 μL 5mg/mL MTT reagent (Sigma) was added for 4 h. A total of 100 μL dimethylsulfoxide (DMSO; Sangon Biotech) was added, and the 96-well plates were shaken to dissolve the formazan product. The absorbance (570 nm) was measured using the auto-microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Transwell Migration and Invasion Assays
Transwell migration assay was conducted using Transwell pore polycarbonate membrane insert (Corning Incorporation, Corning, NY, USA). NSCLC cells were seeded into the upper chambers and cultured in serum-free medium, while the lower chambers were filled with culture medium added with 20% FBS (Gibco). NSCLC cells migrated to the lower surface were immobilized using methanol (Sangon Biotech) and stained using crystal violet (Sangon Biotech). Magnification time: 100 ×.
Transwell invasion assay was carried out using Matrigel-coated chambers (Corning Incorporation) similar to transwell migration assay. Magnification time: 100 ×.
Cell Apoptosis Analysis
NSCLC cells were digested using 0.25% trypsin, and PBS (Sangon Biotech) was used to wash these cells. Cell apoptotic rate was analyzed using Annexin V-fluorescein isothiocyanate (FITC)/PI staining Kit (BD Biosciences, Franklin Lakes, NJ, USA). Annexin V-FITC (5 μL) and PI (5 μL) were added to simultaneously stain these cells. 1 × 104 NSCLC cells were analyzed via flow cytometry.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Complementary DNA (cDNA) was synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo, Tokyo, Japan) or miRNA reverse transcription PCR kit (GenePharma, Shanghai, China). PCR reaction was conducted using the SYBR Premix Ex Taq II Kit (Takara, Tokyo, Japan). The relative expression of circ_SATB2, SATB2, miR-33a-5p and E2F7 was analyzed by the 2−ΔΔCt method and then normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6. Primer sequences were shown in Table 1.
 |
Table 1 Primers in qRT-PCR |
RNase R Digestion
Total RNA samples (2 μg) were incubated with RNase R (6 U; Epicentre Technologies, Madison, WI, USA) for 30 min at room temperature. qRT-PCR was implemented for the determination of circ_SATB2 and SATB2 expression.
Cell Transfection
Recombinant plasmid overexpressing circ_SATB2 (circ_SATB2; Ribobio, Guangzhou, China) and empty vector pLCDH-cir (Vector; Ribobio), recombinant lentiviral plasmid stably overexpressing circ_SATB2 (lenti-circ_SATB2; Ribobio) and corresponding control (lenti-NC; Ribobio), E2F7 small interfering RNA (si-E2F7; GenePharma) and corresponding negative control siRNA (si-NC; GenePharma), miR-33a-5p mimics (miR-33a-5p; GenePharma) and matching control miRNA (miR-NC; GenePharma), miR-33a-5p inhibitor (anti-miR-33a-5p; GenePharma) and matching control (anti-miR-NC; GenePharma) were transfected into NSCLC cells in logarithmic growth phase using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA).
Western Blot Assay
NSCLC cells were harvested using cold PBS buffer (Sangon Biotech). Radioimmunoprecipitation assay buffer (RIPA; Invitrogen) plus protease inhibitor cocktail was used to disrupt the NSCLC cells. Protein supernatant in each sample was removed into the new tube following high-speed centrifugation. Protein samples were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA). After blocking using 5% W/V skimmed milk, the membrane was incubated with the primary antibodies overnight at 4°C, including anti-B cell leukemia/lymphoma 2 (anti-Bcl-2; ab32124; Abcam, Cambridge, MA, USA), anti-BCL2 associated X, apoptosis regulator (anti-Bax; ab32503; Abcam), anti-Cleaved poly(ADP-ribose) polymerase (anti-Cleaved PARP; ab32064; Abcam), anti-E2F7 (ab56022; Abcam) and anti-GAPDH (ab8245; Abcam). Subsequently, the membrane was incubated with the matching horseradish-peroxidase (HRP)-labeled secondary antibody (Abcam). The protein bands were visualized by the enhanced chemiluminescence (ECL) Detection System (GE Healthcare, Chicago, IL, USA).
Bioinformatic Analysis
To investigate the interacted miRNAs with circ_SATB2 or the interacted mRNAs with miR-33a-5p, StarBase software was used to analyze the complementary sites between the target molecule and possible candidate molecules.
Dual-Luciferase Reporter Assay
The partial fragment in circ_SATB2 or the 3ʹ untranslated region (3ʹUTR) of E2F7, containing the wild-type matching sites with miR-33a-5p, was amplified and cloned into pmirGLO vector (Promega, Madison, WI, USA) to generate circ_SATB2-WT or E2F7-WT using SacI (Takara) and XhoI (Takara) enzymes. Furthermore, the complementary sequence in circ_SATB2 or the 3ʹUTR of E2F7 was altered and inserted into the pmirGLO vector (Promega) to obtain circ_SATB2-MUT or E2F7-MUT as the control. A549 and H460 cells in 12-well plates were co-transfected with miR-NC or miR-33a-5p and luciferase plasmids. The activities of Firefly and Renilla were measured with the luminescent detection system (Tecan Group, Mannedorf, Switzerland). The relative luciferase activity was analyzed by using Renilla luciferase activity as the control.
Xenografts in Nude Mice
BALB/c nude mice were purchased from Vital River Laboratory Animal Technology (Beijing, China) with an average weight of about 20 g, and these nude mice were arbitrarily divided into 4 groups, 5 mice in each group. A549 cells stably expressing circ_SATB2 or control A549 cells (2 × 106 cells suspended using 200 μL PBS) were injected into the mice. Nude mice in Cela treatment group were treated with Cela (2 mg/kg) for 4 times per week. Tumor volume was monitored every 5 d as length×width2×0.5. Tumors were dissected after 32 d-injection, and the weight of tumors was recorded. The expression of circ_SATB2, miR-33a-5p and E2F7 mRNA and protein was measured by qRT-PCR or Western blot assay. All protocols in animal experiments were approved by the Committee on the Ethics of Animal Experiments of The Central Hospital of Enshi Tujia And Miao Autonomous Prefecture. All animal procedures were conducted following the Guidelines for Care and Use of Laboratory Animals of “National Institutes of Health”.
Statistical Analysis
GraphPad Prism 7.0 was used for statistical analysis. Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test was used to compare variables in two groups or multiple groups. The data were represented as mean ± standard deviation (SD). Linear correlation was analyzed by Spearman correlation coefficient. P value of less than 0.05 was considered to indicate the statistically significant difference.
Results
Cela Stimulation Suppresses Cell Proliferation, Migration, Invasion and Triggers Cell Apoptosis in NSCLC Cells
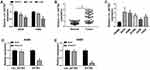
A549 and H460 cells were stimulated with different doses (1 μM or 3 μM) of Cela for 24 h to investigate the biological influences of Cela in the malignant phenotypes of NSCLC cells. Cell proliferation was analyzed by flow cytometry, colony formation assay and MTT assay. After 3 μM Cela treatment, cell number in G0/G1 phase was notably increased, while the number of NSCLC cells in S phase was significantly decreased (Figure 1A and B), suggested that a high dose of Cela suppressed cell cycle progression of NSCLC cells. The number of visible colonies was decreased with the stimulation of 3 μM Cela compared with control group or 1 μM Cela treatment group (Figure 1C), which demonstrated that the proliferation ability of NSCLC cells was restrained after 3 μM Cela stimulation. Through analyzing the cell proliferation curve via MTT assay, we found that cell proliferation was blocked with 3 μM Cela treatment (Figure 1D and E). Subsequently, cell migration, invasion and apoptosis were analyzed by transwell migration assay, transwell invasion assay and flow cytometry. Both the numbers of migrated and invaded NSCLC cells were reduced with 3 μM Cela treatment (Figure 1F and G), suggested that 3 μM Cela suppressed the migration and invasion abilities of NSCLC cells. Cell apoptosis of NSCLC cells was triggered with Cela treatment, especially in 3 μM Cela treatment group (Figure 1H). These results together demonstrated that Cela restrained the proliferation and metastasis while induced the apoptosis of NSCLC cells.
Circ_SATB2 is Highly Expressed in NSCLC Tissues and Cell Lines
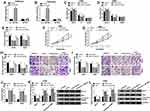
Circ_SATB2 expression was decreased in NSCLC cells after 3 μM Cela treatment (Figure 2A). The expression profile of circ_SATB2 in NSCLC was explored. A total of 49 pairs of NSCLC tumor tissues along with adjacent normal tissues were collected for the determination of circ_SATB2 expression. Compared with adjacent normal tissues, circ_SATB2 abundance was significantly enhanced in NSCLC tumor tissues (Figure 2B). The expression of circ_SATB2 was analyzed in 16HBE and five lung cancer cell lines (A549, H460, H1299, H226 and H522). Circ_SATB2 expression was elevated in all five lung cancer cell lines when compared with human bronchial epithelioid cell line 16HBE (Figure 2C), and A549 and H460 cell lines were chosen for the following experiments due to their higher expression of circ_SATB2 than the other three lung cancer cell lines. CircRNAs are characterized by covalently closed circular structure without 5ʹ or 3ʹ end. We tested if circ_SATB2 was resistant to RNase R to verify its stability, and its matching linear counterpart (SATB2 mRNA) was used as the control. Circ_SATB2 expression was unaffected with or without RNase R digestion, while SATB2 mRNA level was significantly reduced after RNase R digestion compared with Mock group (Figure 2D and E). These findings suggested that circ_SATB2 was aberrantly expressed in NSCLC, and it might be an important regulator for NSCLC progression.
Cela Suppresses NSCLC Progression Through Reducing circ_SATB2 Expression
To investigate the role of circ_SATB2 in Cela-treated NSCLC cells, we treated A549 and H460 cells with Cela (3 μM), Cela (3 μM) + Vector or Cela (3 μM) + circ_SATB2. Prior to functional experiments, we firstly measured the overexpression efficiency of circ_SATB2 plasmid in NSCLC cells. Circ_SATB2 plasmid transfection markedly increased the expression of circ_SATB2 in Cela-induced NSCLC cells, while SATB2 mRNA level was unaffected in two groups (Figure 3A and B). The distribution of NSCLC cells in G0/G1 phase was reduced while there was an increased percentage of NSCLC cells in S phase in circ_SATB2 transfected group in Cela-induced NSCLC cells (Figure 3C and D). The number of colonies was reduced after Cela stimulation, and the transfection of circ_SATB2 partly regained the number of colonies (Figure 3E). Cela stimulation-mediated inhibitory effect on cell proliferation was partly overturned by the addition of circ_SATB2 overexpression plasmid via MTT assay (Figure 3F and G). The results of flow cytometry, colony formation assay and MTT assay together demonstrated that Cela treatment impeded cell proliferation ability, and circ_SATB2 overexpression recovered the proliferation ability in Cela-induced NSCLC cells. Migrated cell number was reduced after Cela treatment, and circ_SATB2 transfection largely rescued the number of migrated NSCLC cells via transwell migration assay (Figure 3H). Meanwhile, the invaded cell numbers in four groups revealed a similar phenomenon to the migrated cell numbers (Figure 3I). The results of transwell assays suggested that Cela-mediated suppression in cell migration and invasion was partly overturned by circ_SATB2 overexpression. Cell apoptosis was analyzed through counting apoptotic rate and measuring the expression of pro-apoptotic proteins (Bax and Cleaved PARP) and anti-apoptotic protein Bcl-2 via flow cytometry and Western blot assay. Cela-induced cell apoptosis was partly attenuated by circ_SATB2 transfection (Figure 3J). The abundance of pro-apoptotic proteins (Bax and Cleaved PARP) was elevated by Cela treatment, and their levels were reduced in Cela and circ_SATB2 co-treated group (Figure 3K and L). Besides, Bcl-2 expression was reduced with Cela treatment, and circ_SATB2 transfection recovered its expression in NSCLC cells (Figure 3K and L). Taken together, Cela-mediated suppression on cell proliferation and metastasis and promotion in cell apoptosis were based on the reduction of circ_SATB2 in NSCLC cells.
Circ_SATB2 Overexpression Alleviates Cela-Induced Suppressive Effect in NSCLC Progression Through Up-Regulating E2F7
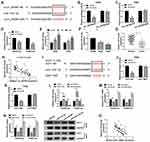
E2F7 mRNA level was found to be significantly up-regulated in NSCLC tumor tissues (n=49) in comparison with that in adjacent non-tumor tissues (n=49) (Figure 4A). Given the results that circ_SATB2 was up-regulated in NSCLC tissues, we analyzed if there was a positive correlation between the expression of circ_SATB2 and E2F7. As displayed in Figure 4B, there was a positive linear correlation between the expression of circ_SATB2 and E2F7. The results of Western blot assay suggested that E2F7 protein level was also increased in NSCLC tumor specimens compared with normal specimens (Figure 4C). The expression of E2F7 mRNA and protein was reduced in 1 μM and 3 μM Cela treatment group compared with Control group, especially in 3 μM Cela treatment group (Figure 4D and E). The levels of E2F7 mRNA and protein were both up-regulated in two NSCLC cell lines compared with 16HBE cell line (Figure 4F and G). To test if circ_SATB2 functioned in Cela-induced NSCLC cells through regulating E2F7, we conducted rescue experiments through treating A549 and H460 cells with Vector + Cela, circ_SATB2 + Cela, circ_SATB2 + si-NC + Cela or circ_SATB2 + si-E2F7 + Cela. E2F7 mRNA and protein enrichment was elevated with the overexpression of circ_SATB2, and the co-transfection with si-E2F7 decreased the mRNA and protein abundance of E2F7 (Figure 4H and I). Circ_SATB2 overexpression-mediated promotion in cell cycle progression in Cela-treated NSCLC cells was partly counteracted by the silencing of E2F7 (Figure 4J and K). E2F7 knockdown counteracted the promoting effect of circ_SATB2 overexpression on the colony formation ability of Cela-treated NSCLC cells (Figure 4L). MTT assay revealed that circ_SATB2 overexpression-mediated promotion in cell proliferation was alleviated by the silencing of E2F7 in Cela-treated NSCLC cells (Figure 4M and N). The motility of NSCLC cells was promoted with the overexpression of circ_SATB2 in Cela-stimulated NSCLC cells, and cell migration and invasion were restrained in Cela + circ_SATB2 + si-E2F7 group (Figure 4O and P). The apoptotic rate was suppressed with the accumulation of circ_SATB2, and the introduction of si-E2F7 increased the apoptotic rate again (Figure 4Q). Circ_SATB2 overexpression decreased the expression of Bax and Cleaved PARP while up-regulated the level of Bcl-2, and these effects were partly overturned by the addition of si-E2F7 (Figure 4R and S). These results suggested that circ_SATB2 counteracted Cela-induced effects in NSCLC cells by enhancing the expression of E2F7.
MiR-33a-5p is a Target of circ_SATB2, and miR-33a-5p Also Binds to the 3ʹUTR of E2F7 in NSCLC Cells
We conducted a bioinformatic analysis using StarBase software to explore the underlying mechanism. MiR-33a-5p was predicted as one of the targets of circ_SATB2 via StarBase software, and the complementary sites along with the mutant sites with miR-33a-5p in circ_SATB2 were shown in Figure 5A. In circ_SATB2-WT transfected group, compared with miR-NC addition group, the transfection of miR-33a-5p resulted in a significant reduction in luciferase activity (Figure 5B and C), suggested that miR-33a-5p bound to circ_SATB2 in NSCLC cells. Furthermore, we constructed luciferase mutant plasmid circ_SATB2-MUT to confirm the binding sites between circ_SATB2 and miR-33a-5p. In circ_SATB2-MUT group, there was no significant comparison in luciferase activity with the co-transfection of miR-NC or miR-33a-5p (Figure 5B and C), suggested that circ_SATB2 bound to miR-33a-5p via it “CAAUGCA” sequence in NSCLC cells. After overexpressing circ_SATB2 in Cela-stimulated NSCLC cells, miR-33a-5p expression was markedly reduced (Figure 5D). With the increased doses of Cela, the expression of miR-33a-5p was gradually up-regulated in NSCLC cells (Figure 5E). The abundance of miR-33a-5p was reduced in NSCLC cell lines and tissues compared with 16HBE cell line and normal tissues (Figure 5F and G). MiR-33a-5p expression was negatively correlated with the level of circ_SATB2 (Figure 5H). The interaction between E2F7 and miR-33a-5p was also predicted by StarBase software and the binding sequence is shown in Figure 5I. Compared with miR-NC and E2F7-WT co-transfected group, miR-33a-5p transfection significantly reduced the luciferase activity in E2F7-WT group (Figure 5J and K), suggesting the target relationship between E2F7 and miR-33a-5p. However, luciferase activity remained unchanged in E2F7-MUT group with the co-transfection of miR-NC or miR-33a-5p (Figure 5J and K), suggested that the sequence “AAUGCA” in E2F7 was the only binding sequence between miR-33a-5p and E2F7 in NSCLC cells. MiR-33a-5p overexpression resulted in a notable up-regulation in miR-33a-5p expression, and miR-33a-5p level was decreased with the transfection of miR-33a-5p inhibitor (anti-miR-33a-5p) in Cela-treated NSCLC cells (Figure 5L). MiR-33a-5p overexpression reduced the mRNA and protein expression of E2F7, and miR-33a-5p silencing caused a notable up-regulation in E2F7 mRNA and protein expression in Cela-induced NSCLC cells (Figure 5M and N). There was a negative correlation between miR-33a-5p and E2F7 mRNA expression in NSCLC tissues (Figure 5O). Overall, there was targeted and negative interaction between miR-33a-5p and circ_SATB2 or E2F7 in NSCLC cells.
Circ_SATB2 Attenuates Cela-Induced Suppressive Influences in the Malignant Behaviors of NSCLC Cells Through Sponging miR-33a-5p
A549 and H460 cells were treated with Cela, Cela + Vector, Cela + circ_SATB2, Cela + circ_SATB2 + miR-NC or Cela + circ_SATB2 + miR-33a-5p. The expression of miR-33a-5p was reduced in Cela and circ_SATB2 group, while miR-33a-5p overexpression recovered its expression in NSCLC cells (Figure 6A). Cela-induced reduction in E2F7 mRNA and protein expression was counteracted by the overexpression of circ_SATB2, and the mRNA and protein expression of E2F7 was decreased with the addition of miR-33a-5p (Figure 6B and C), suggested that E2F7 was regulated by circ_SATB2/miR-33a-5p axis in Cela-induced NSCLC cells. Cell cycle progression was accelerated with the overexpression of circ_SATB2 in Cela-induced NSCLC cells, and the addition of miR-33a-5p suppressed the cell cycle progression in NSCLC cells treated with Cela (Figure 6D and E). The results of colony formation assay and MTT assay demonstrated that circ_SATB2-mediated promotion in cell proliferation in Cela-treated NSCLC cells was partly overturned by the introduction of miR-33a-5p (Figure 6F–H). Cell metastasis was accelerated with the overexpression of circ_SATB2, and miR-33a-5p addition suppressed the migration and invasion of Cela-induced NSCLC cells again (Figure 6I and J). Circ_SATB2 overexpression-mediated suppressive effect in cell apoptosis was reversed by the addition of miR-33a-5p mimic in Cela-induced NSCLC cells (Figure 6K). Circ_SATB2 overexpression reduced the levels of Bax and Cleaved PARP while increased the level of Bcl-2, and these influences were attenuated by the addition of miR-33a-5p in NSCLC cells stimulated by Cela (Figure 6L and M). These findings demonstrated that circ_SATB2 accelerated NSCLC progression through sponging miR-33a-5p in Cela-treated NSCLC cells.
Cela-Mediated Inhibitory Influence in the Growth of NSCLC Tumors is Overturned by the Accumulation of circ_SATB2 in vivo
To confirm the roles of Cela and circ_SATB2 in the progression of NSCLC in vivo, we conducted tumor xenograft assay. Cela treatment suppressed the growth of NSCLC tumors by analyzing the tumor growth curve and the weight of dissected tumors (Figure 7A and B). Furthermore, the suppressive effect of Cela was attenuated by circ_SATB2 overexpression (Figure 7A and B), suggested that Cela suppressed NSCLC tumor growth by decreasing the abundance of circ_SATB2 in vivo. The expression of circ_SATB2 and E2F7 mRNA and protein were decreased in Cela treatment group, and the overexpression of circ_SATB2 recovered the expression of these molecules (Figure 7C, E and F). Cela treatment up-regulated miR-33a-5p expression, and miR-33a-5p expression was reduced in Cela and lenti-circ_SATB2 group (Figure 7D). These results demonstrated that Cela suppressed NSCLC tumor growth partly through down-regulating circ_SATB2 in vivo.
 |
Figure 7 Cela-mediated inhibitory influence in the growth of NSCLC tumors is overturned by the accumulation of circ_SATB2 in vivo. (A) Tumor volume in four groups was monitored every 5 d after injection for 12 d to generate tumor growth curve. (B) Tumors were dissected from mice after injection for 32 d, and the tumor weight was calculated as length×width2×0.5. (C and D) The expression of circ_SATB2 and miR-33a-5p in tumor tissues was examined by qRT-PCR. (E and F) The mRNA and protein levels of E2F7 in tumor tissues were determined by qRT-PCR and Western blot assay. *P<0.05. |
Discussion
High metastasis and drug resistance remain two obstacles for NSCLC therapy. Therefore, identifying novel targets and effective drugs for NSCLC treatment are urgently needed. Natural compounds are crucial sources for novel therapeutic drugs.22 Cela is extracted from Tripterygium wilfordii, and it exerts anti-tumor activity in various cancers, including NSCLC.8,23 Mou et al demonstrated that Cela triggered the apoptosis of NSCLC cells.23 Nevertheless, the working mechanism of Cela in NSCLC progression has not been fully understood. We found that Cela exerted the anti-tumor activity in NSCLC through targeting circ_SATB2/miR-33a-5p/E2F7 signaling cascade.
Cela stimulation blocked cell cycle progression, colony formation ability, proliferation and metastasis and triggered cell apoptosis of NSCLC cells. Circ_SATB2 expression was significantly reduced after Cela treatment. CircRNAs are known to regulate the progression of many cancers, including NSCLC. For instance, circ_0067934 knockdown restrained the proliferation, migration and invasion of NSCLC cells, and high expression of circ_0067934 was associated with the dismal prognosis of NSCLC patients.24 Circ_ZNF124 contributed to NSCLC progression by targeting miR-337-3p/JAK2/STAT3 signaling.25 Zhang et al found that circ_SATB2 accelerated NSCLC progression through acting as miR-326 sponge to up-regulate FSCN1 expression.15 Here, we found that circ_SATB2 was highly expressed in NSCLC tissues and cell lines. Subsequently, we investigated if Cela suppressed NSCLC progression by reducing circ_SATB2 expression. Cela stimulation-mediated suppressive influences in the malignant behaviors of NSCLC cells were all attenuated by the overexpression of circ_SATB2, suggested that circ_SATB2 was an important downstream molecule by which Cela exerted its anti-tumor role.
The working mechanisms of circRNAs in cancers are multiple. CircRNAs could function as miRNA sponges to influence the levels of miRNA target genes.26,27 Through using bioinformatic software StarBase, we found that circ_SATB2 had many miRNA-binding sites, demonstrated that circ_SATB2 might exert its oncogenic role through acting as miRNA sponges. MiR-33a-5p was the target of circ_SATB2, and it was negatively regulated by circ_SATB2. Cela stimulation markedly up-regulated miR-33a-5p expression in NSCLC cells. As a tumor suppressor, miR-33a-5p was reported to be associated with the progression of many cancers. Yan et al found that miR-33a-5p restrained the development of colorectal cancer through targeting and suppressing MTHFD2.28 Guan et al found that miR-33a-5p accumulation elevated the chemosensitivity of triple-negative breast cancer cells to doxorubicin.29 Pan et al demonstrated that miR-33a-5p and miR-128-3p were both down-regulated in lung cancer, and the expression of miR-33a-5p and miR-128-3p in whole blood might be novel markers for the early diagnosis of lung cancer.30 Wang et al claimed that circCCDC66 accelerated the proliferation, migration and invasion while suppressed the apoptosis of NSCLC cells through targeting miR-33a-5p/KPNA4 axis.17 Circ_SATB2 overexpressing-mediated influences in Cela-treated NSCLC cells were alleviated by the addition of miR-33a-5p, suggested that circ_SATB2 counteracted Cela-mediated suppressive effect in NSCLC progression through down-regulating miR-33a-5p.
E2F7 is a member of E2F family, and E2F7 acts as a transcription repressor to function. Accumulating works demonstrated that E2F7 functioned as an initiator to modulate cell cycle progression, cell apoptosis and differentiation.19,31,32 Liu et al found that E2F7 expression was elevated in estrogen receptor-positive breast cancer, and the down-regulation of miR-26a and the elevation in E2F7 expression elevated TAM resistance in MCF-7 cells.33 Wang et al demonstrated that E2F7 abundance was enhanced in NSCLC tissues, and miR-935 restrained NSCLC progression through targeting and reducing E2F7 expression.21 E2F7 was confirmed as a target of miR-33a-5p in NSCLC cells, and E2F7 silencing overturned circ_SATB2-mediated effects in Cela-treated NSCLC cells, suggested that circ_SATB2 functioned in Cela-stimulated NSCLC cells through enhancing E2F7 expression. However, the potential mechanism by which E2F7 contributed to the malignant behaviors of NSCLC cells remains to be clarified. E2F7 was involved in the regulation of tumor metastasis and development through modulating the process of epithelial–mesenchymal transition (EMT) and PI3K/AKT signaling.34,35 Wang et al found that miR-935 restrained the motility and proliferation of NSCLC cells through reducing E2F7 expression and inactivating AKT signaling.21 In future, the downstream pathway behind E2F7-induced progression in NSCLC needs to be deeply investigated. The regulatory relationship among these three molecules was explored. Circ_SATB2 enhanced E2F7 mRNA and protein expression by targeting miR-33a-5p expression in NSCLC cells. Cela suppressed NSCLC tumor growth, and the suppressive activity of Cela in NSCLC was alleviated by the overexpression of circ_SATB2 in vivo. These findings further suggested that Cela restrained NSCLC progression through reducing circ_SATB2 expression in vivo.
In conclusion, circ_SATB2 counteracted Cela-mediated inhibitory influence in NSCLC progression through targeting miR-33a-5p/E2F7 axis. Circ_SATB2/miR-33a-5p/E2F7 axis may be potential target for NSCLC therapy.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Molina JR, Yang P, Cassivi SD. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594.
2. Hussain S. Nanomedicine for Treatment of Lung Cancer. Adv Exp Med Biol. 2016;890:137–147.
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
4. Reck M, Popat S, Reinmuth N, De Ruysscher D. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25 Suppl 3:iii27–39.
5. Araz O, Ucar EY, Meral M, et al. Frequency of Class I and II HLA alleles in patients with lung cancer according to chemotherapy response and 5-year survival. Clin Respir J. 2015;9(3):297–304.
6. Latimer KM, Mott TF. Lung cancer: diagnosis, treatment principles, and screening. Am Fam Physician. 2015;91(4):250–256.
7. Wang Y, Liu Q, Chen H, et al. Celastrol improves the therapeutic efficacy of EGFR-TKIs for non-small-cell lung cancer by overcoming EGFR T790M drug resistance. Anticancer Drugs. 2018;29(8):748–755.
8. Fan XX, Li N, Wu JL, et al. Celastrol induces apoptosis in gefitinib-resistant non-small cell lung cancer cells via caspases-dependent pathways and Hsp90 client protein degradation. Molecules. 2014;19(3):3508–3522.
9. Lo Iacono M, Monica V, Vavalà T, et al. ATF2 contributes to cisplatin resistance in non-small cell lung cancer and celastrol induces cisplatin resensitization through inhibition of JNK/ATF2 pathway. Int J Cancer. 2015;136(11):2598–2609.
10. Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816.
11. Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
12. Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. doi:10.1093/nar/gkv045
13. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
14. Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503(3):1503–1509. doi:10.1016/j.bbrc.2018.07.070
15. Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19(1):101. doi:10.1186/s12943-020-01221-6
16. Zhang C, Zhang Y, Ding W, Lin Y, Huang Z, Luo Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6(12):881–889. doi:10.1007/s13238-015-0223-8
17. Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019.
18. Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19(6):326–338.
19. Endo-Munoz L, Dahler A, Teakle N, et al. E2F7 can regulate proliferation, differentiation, and apoptotic responses in human keratinocytes: implications for cutaneous squamous cell carcinoma formation. Cancer Res. 2009;69(5):1800–1808. doi:10.1158/0008-5472.CAN-08-2725
20. Reimer D, Sadr S, Wiedemair A, et al. Clinical Relevance of E2F Family Members in Ovarian Cancer--An Evaluation in a Training Set of 77 Patients. Clin Cancer Res. 2015;12(4):144–151. doi:10.1158/1078-0432.CCR-06-0780
21. Wang C, Li S, Xu J, Niu W, Li S. microRNA-935 is reduced in non-small cell lung cancer tissue, is linked to poor outcome, and acts on signal transduction mediator E2F7 and the AKT pathway. Br J Biomed Sci. 2019;76(1):17–23.
22. Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–661.
23. Mou H, Zheng Y, Zhao P, Bao H, Fang W, Xu N. Celastrol induces apoptosis in non-small-cell lung cancer A549 cells through activation of mitochondria- and Fas/FasL-mediated pathways. Toxicol in Vitro. 2011;25(5):1027–1032.
24. Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22(10):3053–3060.
25. Li Q, Huang Q, Cheng S, Wu S. Circ_ZNF124 promotes non-small cell lung cancer progression by abolishing miR-337-3p mediated downregulation of JAK2/STAT3 signaling pathway. Cancer Cell Int. 2019;19:291.
26. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
27. Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842.
28. Yan Y, Zhang D, Lei T, et al. MicroRNA-33a-5p suppresses colorectal cancer cell growth by inhibiting MTHFD2. Clin Exp Pharmacol Physiol. 2019;46(10):928–936.
29. Guan X, Gu S, Yuan M, Zheng X, Wu J. MicroRNA-33a-5p overexpression sensitizes triple-negative breast cancer to doxorubicin by inhibiting eIF5A2 and epithelial-mesenchymal transition. Oncol Lett. 2019;18(6):5986–5994.
30. Pan J, Zhou C, Zhao X, et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci Rep. 2018;8(1):16699.
31. Salvatori B, Iosue I, Mangiavacchi A, et al. The microRNA-26a target E2F7 sustains cell proliferation and inhibits monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis. 2012;3(10):e413.
32. Carvajal LA, Hamard PJ, Tonnessen C, et al. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012;26(14):1533–1545.
33. Liu J, Li X, Wang M, et al. A miR-26a/E2F7 feedback loop contributes to tamoxifen resistance in ER-positive breast cancer. Int J Oncol. 2018;53(4):1601–1612.
34. Mahmood MQ, Ward C, Muller HK, et al. Epithelial mesenchymal transition (EMT) and non-small cell lung cancer (NSCLC): a mutual association with airway disease. Med Oncol. 2017;34(3):45.
35. Jakubíková J, Sedlák J, Mithen R, et al. Role of PI3K/Akt and MEK/ERK signaling pathways in sulforaphane- and erucin-induced Phase II enzymes and MRP2 transcription, G2/M arrest and cell death in Caco-2 cells. Biochem Pharmacol. 2005;69(11):1543–1552.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.