Back to Journals » OncoTargets and Therapy » Volume 13
CircRNAs: A New Chapter in Oral Squamous Cell Carcinoma Biology
Authors Fan H, Jiang J, Tang Y, Liang X, Tang Y
Received 22 May 2020
Accepted for publication 16 August 2020
Published 11 September 2020 Volume 2020:13 Pages 9071—9083
DOI https://doi.org/10.2147/OTT.S263655
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Hua-yang Fan,1,* Jian Jiang,2,* Ya-jie Tang,3 Xin-hua Liang,1 Ya-ling Tang1
1State Key Laboratory of Oral Diseases and National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, Sichuan, People’s Republic of China; 2Department of Head and Neck Surgery, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, School of Medicine, University of Electronic Science and Technology of China, Chengdu, Sichuan, People’s Republic of China; 3State Key Laboratory of Microbial Technology, Shandong University, Qingdao, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xin-hua Liang; Ya-ling Tang Email [email protected]; [email protected]
Abstract: With the rapid development of bioinformatics and gene sequencing technologies, understanding of circular RNAs (circRNAs) has been extended, and numerous studies have identified the key regulator role of circRNAs in a variety of diseases, especially in cancer. Recently, accumulated studies of oral squamous cell carcinoma (OSCC) have discovered the great potential of circRNAs, which can serve as prognostic or diagnostic biomarkers and affect the development and therapy of OSCC. In this review, we detail the new progress of circRNA research for OSCC in order to provide new strategies for clinical diagnosis and treatment.
Keywords: circular RNA, oral squamous cell carcinoma, miRNA sponge, biomarker
Introduction
Oral squamous cell carcinoma (OSCC) includes cancers that occur in the mouth and oropharynx, accounting for about 90% of all oral malignancies.1 Worldwide, the incidence of OSCC is approximately 4/100,000, more common in men and the elderly.2 In some Asia-Pacific countries, its incidence ranks among the top three of all cancers.3 Moreover, they are usually diagnosed at an advanced stage, and environmental risk factors, viral infections, as well as genetic changes all augment the incidence of OSCC. Currently, the 5-year overall survival rate of OSCC patients is estimated to be 50% to 60%.4 Therefore, it is necessary to clarify the molecular mechanisms of OSCC pathogenesis, which will help to develop more effective diagnostic methods and treatment strategies.
Circular RNA (circRNA) is a subclass of non-coding RNA (ncRNA). After long non-coding RNA (lncRNA) and microRNA (miRNA), it has developed into a new research hotspot in the field of cancer.5 CircRNAs are produced by reverse splicing and are characterized by a closed single-stranded structure and lack of 5 ‘cap and 3ʹ polyadenylation (poly(A)) tail, which makes them more stable than lncRNA and miRNA.6 And circRNAs are highly conserved in eukaryotes, and their expression exhibits tissue- and developmental stage-specific. Numerous studies have revealed that circRNAs have an important effect on the progression and treatment of cancer and play a regulatory role in the tumor microenvironment (TME). Therefore, it suggests that circRNAs may serve as new cancer biomarkers as well as potential therapy targets.7–9
At present, plenty of studies have been devoted to exploring the function of circRNAs in OSCC and found that copious abnormally expressed circRNAs are closely related to the clinical characteristics of OSCC, indicating that circRNAs can be used as biomarkers for OSCC. In addition, aberrantly expressed circRNAs can act as miRNA sponges or participate in several cancer-related signaling pathways, thereby altering the progression of OSCC and affecting drug resistance and radiotherapy in OSCC patients. In this review, we comprehensively discuss the research progress of circRNAs in OSCC and outline their functions.
Characterization and Main Functions of CircRNA
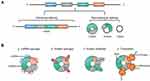
CircRNAs are produced by a non-canonical splicing event named back-splicing, during such event the downstream splice-donor site is covalently linked to upstream splice-acceptor site.10 The formed closed-loop structure of circRNAs makes them much more stable than linear RNAs, and their half-life is about 4 times longer than linear RNAs.11 In addition, back-splicing makes circRNAs lack 5’ cap and 3ʹ tail, which enables them resistant to ribonuclease RNase R. This feature is usually used in experiments to distinguish circRNAs from linear counterparts.12 According to the sequences it contains, circRNA can be divided into three main different types: exon circRNA (EcRNA), intron circRNA (CiRNA) and exon-intron circRNA (EIcRNA).13 Among them, EcRNA is the most common and is mainly distributed in the cytoplasm, while circRNAs containing introns are distributed in the nucleus.14 CircRNAs exhibit widespread expression in different species and abundant expression in human tissues, especially in brain.15 And their sequences are highly conserved, and many studies have detected about 15,000 human circRNAs sequences in mice.15,16 Moreover, their expression exhibits in tissue-, cell- and developmental stage-specific manner, and partially dysregulated circRNAs have a close relationship with tumor pathological differentiation, TNM stage and Lei et al, indicating that they may be suitable candidates for cancer biomarkers.17–19
CircRNAs have shown multiple functions in a variety of cancers. Currently, the most established function of circRNAs is that circRNAs can act as miRNA sponges, competitively inhibit gene translation and regulate relevant signaling pathways, thereby affecting the progression of cancer.20 Some circRNAs containing RNA binding proteins (RBPs) motifs can serve as protein sponges to indirectly regulate the function of RBPs.21 And a few circRNAs can directly interact with proteins and enzymes to regulate various physiological processes, which is called protein scaffolds.22 Intriguingly, as non-coding RNAs, a handful of circRNAs have been confirmed with protein-coding ability, and currently, the widely accepted mechanism is internal ribosome entry site (IRES)-mediated translation, which is a cap-independent translation mechanism. IRES is a sequence located in the 5ʹ untranslated region (UTR) and can recruit ribosomes to initiate translation, and plenty of IRES sequences have been identified in circRNAs. Several circRNAs have been found to code proteins in human cancers, such as glioma and glioblastoma.23,24 However, the current research is still in the initial stage, and the function of proteins encoded by circRNAs needs further exploration, especially in OSCC (Figure 1).25,26
The fast development of bioinformatics has accelerated the exploration of circRNAs. At present, high-throughput RNA-sequencing and microarray analysis are the most popular detection methods. With the help of find-circ, CIRI2 and other computer algorithms, the unique back-splice junction (BSJ) of each circRNA can be determined, which greatly improves the reliability of detection. And the experimental validation methods are more mature. The combination of RNase R digestion, qRT-PCR based on divergent primers and Northern blotting can accurately verify circRNAs. Furthermore, we can better predict the function of circRNAs based on the establishment of the circRNA databases and sequencing results. Whether they have miRNAs sponges, protein sponges, translation ability or other potentials can be well predicted.
Currently, lncRNAs, which are defined as ncRNAs of more than 200 nucleotides, have been systematically discussed in the OSCC, and they can act as miRNA sponges and participate in cancer-related signaling pathways.27 LncRNAs can also be used as biomarkers, but their stability and conservation are poor. MiRNAs refer to a class of short ncRNAs with a length of about 20 nucleotides, and they played an important role in the development and treatment of OSCC.28 And miRNAs are identified as key molecules in various signaling pathways through miRNA sponge function. Like lncRNAs and miRNAs, circRNAs are also abundantly expressed in body fluids (such as blood and saliva) and tumor tissues in a cancer-specific manner. These ncRNAs are stable in body fluids and their levels are often related to the clinical and pathological features of cancer. They all are inspiring for the early diagnosis of OSCC in a non-invasive way. Moreover, circRNAs with a covalent closed loop structure have higher stability and longer half-life than lncRNAs and miRNAs. And circRNAs exhibited high conservation across different species. Extensive research on the association of lncRNAs-miRNAs and circRNAs-miRNAs may help to elucidate the complex pathological mechanism of OSCC.
CircRNAs in OSCC
The function of circRNAs in the development of various cancers has been extensively studied. In recent years, research on circRNA in the field of OSCC has been increasing, and it was found that circRNAs have an important impact on the progression, treatment and prognosis of OSCC (Supplementary Figure 1).
Expression of CircRNAs in OSCC
Ascending evidence reveals that the abnormal expression of circRNAs is related to the development and prognosis of OSCC. CircPVT1 was overexpressed in OSCC patients and its expression was related to mutant p53 protein status in OSCC patients.29 In vitro experiments confirmed that p53 protein depletion can downregulate the expression of circPVT1, but there was no inverse regulation between p53 and circPVT1. In addition, RNA immunoprecipitation (RIP) and chromatin immunoprecipitation (ChIP) experiments showed that circPVT1 did not directly bind to p53 but was regulated by the mut-p53/YAP (Yes-Associated Protein)/TEAD complex. Particularly, circPVT1 can control and enhance its expression in the nucleus. Through high-throughput sequencing and microarray analysis, Wang et al analyzed the expression of circRNAs in 8 pairs of OSCC tissues and controls, with a total of 1921 existing circRNAs and 10,021 new circRNAs.30 Most circRNAs were derived from exons and were distributed on chromosomes 1 and 2. There were 16 circRNAs with significantly different expressions in OSCC and adjacent tissues, with fold changed ≥2.0. Among them, 8 circRNAs were upregulated and 8 circRNAs were downregulated. Deng et al performed high-throughput circRNA microarray analysis on 3 pairs of OSCC and matched normal tissues and filtered them with volcanic maps.31 A total of 213 circRNAs with ≥1.5-fold changes were screened out, of which 124 were upregulated and 89 were downregulated. Under GO and Functional categories analysis, 20 differentially expressed circRNAs were involved in MAPK and PI3K signaling pathways. Moreover, Qiu et al analyzed the expression of circRNAs in tongue squamous cell carcinoma (TSCC).32 High‐throughput sequencing and RT-PCR were performed, and 12,156 circRNAs were identified in TSCC. The results showed that 314 circRNAs were significantly upregulated, while 8 circRNAs were remarkably reduced by more than 2 times compared with adjacent tissues. Under KEGG pathway analysis, copious circRNAs were associated with tumor signaling pathway protiens such as MAPK.
In particular, circRNAs in saliva of OSCC patients also exhibited aberrant expression. Through microarray screening and qRT-PCR verification, Zhao et al detected 32 abnormally expressed circRNAs in the saliva of OSCC patients, including 12 overexpressed circRNAs and 20 under-expressed circRNAs.33 A large number of experiments have identified the function of these dysregulated circRNAs, showing that the abnormal expression of circRNAs has multiple biological effects. However, it is not clear which circRNAs play a more important role and what is the relation between these circRNAs in the development of OSCC.
Biological Functions of CircRNAs in OSCC
Potential Novel Biomarkers
Dysregulated non-coding (ncRNAs) RNAs in OSCC, including miRNAs and lncRNAs, have been identified as ideal biomarkers.27,34 Similarly, numerous studies have found that circRNAs have great potential as OSCC biomarkers (Table 1).
 |
Table 1 The Potential CircRNA Biomarkers for OSCC |
In OSCC patients with mutant p53 protein, circPVT1 was significantly upregulated.29 And patients with overexpressed circPVT1 usually had a poor survival, suggesting that circPVT1 could be a prognostic biomarker for OSCC. Aberrant circRNAs in OSCC patients were also related to their pathological differentiation and might act as diagnostic biomarkers. For example, hsa_circ_0008309,35 hsa_circ_009755,36 circRNA_000334 and circRNA_00674030 were downregulated in OSCC tissues, and their expression was closely connected with the pathological differentiation of OSCC. Receiver operating characteristic (ROC) curve analysis was performed in hsa_circ_0008309 and hsa_circ_009755, and the area under the curve (AUC) values of hsa_circ_0008309 and hsa_circ_009755 were 0.764 and 0.782, respectively. These results indicated that hsa_circ_0008309, hsa_circ_009755, circRNA_000334, and circRNA_006740 can be used as potential diagnostic biomarkers for OSCC.
Furthermore, circRNAs were also found to be related to TNM stage and tumor size of OSCC. Hsa_circ_00124237 and hsa_circ_007238738 were downregulated in OSCC. The expression of hsa_circ_001242 was significantly connected to tumor size and T stage of OSCC and the expression of hsa_circ_0072387 was correlated to the TNM stage. The AUC values of hsa_circ_001242 and hsa_circ_0072387 were 0.784 and 0.746, respectively. Meanwhile, hsa_circ_010929139 was overexpressed (approximately 4-times) in OSCC patients and was positively associated with their TNM stage. And the prognosis of OSCC patients with upregulated hsa_circ_0109291 was poor. The results indicated that hsa_circ_001242, hsa_circ_0072387 and hsa_circ_0109291 may be biomarkers in OSCC diagnosis and treatment targets. Hsa_circ_0086414,40 hsa_circ_009212541 and circ_000014042 were greatly downregulated in OSCC, and they were notably associated with TNM stage as well as lymph node metastasis. The area below the ROC curve of hsa_circ_0086414 was 0.749. Deng et al screened two circRNAs with the highest and lowest expression in OSCC, hsa_circ_043621 and hsa_circ_102459, respectively.31 These two circRNAs were closely associated with TNM stage, tumor differentiation and lymph node metastasis, suggesting that they can be used as diagnostic biomarkers. Hsa_circ_0001742 was upregulated in TSCC and showed positive correlation with advanced clinical stage and lymph node metastasis.43 Li et al found that another circRNA which was also correlated to lymph node metastasis of OSCC, hsa_circ_0004491, which was greatly reduced in OSCC tissues, and ROC analysis showed its diagnostic value (AUC=0.751) for OSCC.44
CircRNAs differentially expressed in saliva could also serve as biomarkers. Salivary hsa_circ_0001874 and hsa_circ_0001971 were upregulated in OSCC patients.33 It was demonstrated that both hsa_circ_0001874 and hsa_circ_0001971 were correlated to TNM stage, and circ_0001874 was also related to tumor grade. In addition, the AUC of hsa_circ_0001874 and hsa_circ_0001971 of OSCC were 0.863 and 0.845, respectively, and reached 0.922 with their combination. The AUC of their combination for distinguishing OSCC and oral leukoplakia (OLK) was 0.895. After surgery, hsa_circ_0001874 and hsa_circ_0001971 reduced to a level with no significant difference. Therefore, the authors believed that those two circRNAs in saliva might be highly effective biomarkers for early diagnosis of OSCC.
Acting as miRNA Sponges
MicroRNA (miRNA) plays an important role in regulating relevant signaling pathway proteins by base-pairing with mRNA. As one of the competitive endogenous RNA (ceRNA), circRNAs contain miRNA response elements (MREs), which enable circRNAs to interact with target miRNAs and inhibit their activity, thereby affecting mRNA expression. At present, a large number of circRNA-miRNA-mRNA networks have been found to be closely associated with the process of OSCC (Table 2 and Figure 2).
 |
Table 2 CircRNA-miRNA-mRNA Networks and Their Functions in OSCC |
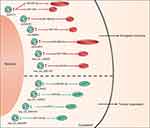 |
Figure 2 Schematic diagram of the circRNA-miRNA networks in OSCC. |
Verduci et al confirmed that circPVT1 could act as a sponge of miR-497-5p, which had anti-tumor effects in a variety of cancers including OSCC.29 The potential downstream carcinogenic effects of circPVT1 was to elevate the expression of aurka, mki67 and bub1 genes, thereby promoting the proliferation of tumor cell. In another study, He et al found that circPVT1 could also effectively sponge miR-125b.45 The circPVT1/miR-125b axis promoted OSCC cell proliferation by increasing the expression of the downstream target STAT3. Circ-PKD2 was downregulated in OSCC patients and it might sponge to miR-204-3p.46 Dual-Luciferase Reporter analysis confirmed the direct interaction between circ-PKD2 and miR-204-3p. By overexpressing circ-PKD2, the carcinogenic ability of miR-204-3p was significantly reduced, resulting in inhibition of OSCC cell proliferation, migration as well as invasion, and induction of cell cycle arrest and apoptosis. In vivo study also showed that overexpression of circ-PKD2 can significantly reduce the size and weight of OSCC xenografted tumor. The downstream signaling pathway was studied, and luciferase reporter gene analysis confirmed that APC2 was the target of miR-204-3p. Circ-PKD2 reduced the inhibitory effect of miR-204-3p and upregulated the expression of APC2, causing the inactivation of β-catenin, protein kinase B and extracellular signal-regulated kinase 1/2 pathway. CircDOCK1 was highly expressed in OSCC cells, and miR-196a-5p was identified as the target of circDOCK1. In addition, BIRC3 was a target of miR-196a-5p, which might cause glioblastoma to escape apoptosis and drug resistance.47 The authors suggested that the circDOCK1/miR-196a-5p/BIRC3 axis played an important role in promoting OSCC cell apoptosis.48 Zhu et al identified hsa_circRNA_100533 as a miRNA sponge of miR‐933 by luciferase reporter assays and RIP, which was downregulated in OSCC and exerted its antitumor function through hsa_circRNA_100533/miR‐933 pathway.49 They also found that GNAS was downregulated in OSCC, which had been identified as a carcinogenic molecule in colorectal cancers and hepatocellular carcinoma.50,51 The luciferase reporter assay confirmed that GNAS was closely related to hsa_circ_100533 and miR-933, and its expression was also regulated by hsa_circ_100533 and miR-933. After silencing GNAS, the inhibitory effect of the hsa_circRNA_100533-miR-933 axis on OSCC was reversed, indicating that the hsa_circ_100533/miR-933/GNAS pathway may play a key role in the process of OSCC. In addition, Xia et al identified a metastasis-associated circRNA in OSCC, hsa_circ_0001162 (circ-MMP9), which was upregulated in OSCC.52 Circ-MMP9 can act as a miR-149 sponge and target AUF1 and has a positive correlation with MMP9 expression. Knocking down circ-MMP can decrease the expression of MMP9, thereby inhibiting the invasion and metastasis of OSCC, which was confirmed in vitro and in vivo. CircUHRF1 was significantly overexpressed in OSCC and could serve as a miR-526b-5p sponge to further up-regulate c-Myc, which can promote the transcription of TGF-β1 and ESRP1. In addition, it was revealed that ESRP1 can assist circUHRF1 circularization and biogenesis, thereby forming a circUHRF1/miR-526b-5p/c-Myc/TGF-β1/ESRP1 feedback loop to promote OSCC tumorigenesis and EMT.53 In vitro experiments, silencing circUHRF1 can repress the proliferation, migration, invasion, as well as EMT abilities of OSCC cells. And in vivo function experiments also revealed that suppression of circUHRF1 can effectively inhibit the growth of OSCC tumors. Chen et al identified that hsa_circ_100290 was significantly upregulated in OSCC and could serve as a sponge for miR‐378a to regulate OSCC cell growth via GLUT1,54 which was the major glucose transporter involved in the glycolysis and was upregulated in OSCC.55 Silencing hsa_circ_100290 significantly increased miR‐378a expression and inhibited OSCC cell proliferation and glycolysis, and overexpressing GLUT1 could rescue this effect. Moreover, miR‐378a can simultaneously bind to hsa_circ_100290 as well as GLUT1and regulate their expression.
CircRNAs in OSCC can function as both biomarkers and miRNA sponges. Peng et al revealed that hsa_circ_0000140 can target and deactivate miR-31, subsequently upregulating LATS2.42 Their study uncovered the role of hsa_circ_0000140/miR-31/LATS2 network in inhibiting OSCC cell tumorigenesis and EMT. In vivo studies, upregulating hsa_circ_0000140 reduced the xenografted tumor growth and lung metastatic by more than 2-fold. In addition, Shao et al proved that hsa_circ_0001742 was a sponge of miR-634, and miR-634 targeted RAB1A. Overexpression of hsa_circ_0001742 could induce TSCC proliferation and invasion via the miR-634/RAB1A axis.43 In another study, hsa_circ_0008309 overexpression downregulated miR-136-5p as well as miR-382-5p and upregulated ATXN1, which was a part of the Notch signaling pathway and was involved in mediating hypoxia-induced migration and invasion. However, more experiments were demanded to prove the role of ATXN1 in OSCC. The author believed that hsa_circ_0008309-miR-136-5P/hsa-miR-382-5P-ATXN might have a regulatory effect on the process of OSCC.35 Li et al demonstrated that hsa_circ_0004491 was decreased in OSCC, and its overexpression can suppress OSCC cell migration and invasion and regulate the expression of EMT-related proteins.44 Based on bioinformatics analysis, they found 3 potential target miRNAs, including hsa-miR-136-5p, hsa-miR-149-5p and hsa-miR-155-5p. After silencing hsa_circRNA_0004491, only the level of has-miR-155-5p changed. SIRT1 was the target of has-miR-155-5p and was involved in the regulation of OSCC proliferation and invasion.56,57 Therefore, the authors predicted that hsa_circ_0004491/miR-155-5p/SIRT1 may affect the procession of OSCC. Moreover, based on bioinformatics analysis, Dou et al found that hsa_circ_0072387 might affect the progression of OSCC through the hsa_circ_0072387/miR-29-29p/miR-141-3p-MMP2/BCL2/PTEN axis but lacked more experimental evidence.38 Similarly, Qiu and his colleagues predicted the miRNA sponge effect of circRNAs and found that each abnormally expressed circRNA in TSCC was associated with 10 tumor-related miRNAs.32
Currently, a large number of circRNAs have been identified as miRNA sponges and exert their important influence in the development of different cancers.58 However, compared with the number of circRNAs that have been identified by sequencing technology, only a handful of circRNAs have been confirmed with biological functions. Therefore, although miRNA sponges are currently the most important biological function of circRNAs, this field is still at an early stage. Moreover, by interacting with miRNA, circRNA can not only act as a sponge to reduce its activity, but also act as a “reservoir” to stabilize and activate miRNA. CiRS-7, the most famous circRNA by far, has more than 70 MREs and can interact with different miRNAs, such as miR-7 and miR-671. CiRS-7 can sponge to miR-7,59 but when ciRS-7 interacts with miR-671 it serves as a miRNA “reservoir” and activates miR-671.60 All above, the research on the miRNA sponge function of circRNA in OSCC has just begun. Their impact on OSCC is still lacking experimental validations, and only few studies have been verified both in vivo and in vitro.
Involved in Multiple Cancer-Associated Signaling Pathways
CircRNA has become an important regulator of carcinogenesis and is involved in various signaling pathways related to the initiation and development of cancer. Dysregulated circRNA in OSCC also affects the proliferation, invasion and metastasis of OSCC by regulating cancer-related signaling pathways, such as MAPK, PI3K/Akt/mTOR, Notch signaling pathways (Table 3).
 |
Table 3 CircRNA-Related Signaling Pathways in OSCC |
Deng et al screened out 2 circRNAs with the highest and lowest expression from 213 dysregulated circRNAs in OSCC, hsa_circ_043621 and hsa_circ_102459, respectively.31 Through GO enrichment and functional category analysis, they had the greatest potential to participate in MAPK and PI3K signaling pathways. It was found that overexpression of hsa_circ_102459 or silence of hsa_circ_043621 can inhibit the proliferation of TSCC1 cells and promote G0/G1 cell cycle arrest and apoptosis. Further research confirmed that hsa_circ_102459 overexpression and circ_043621 knockdown both inhibited MAPK, PI3K/Akt and Bcl-2/BAX axes, thereby inhibiting the progression of OSCC and increasing OSCC cell apoptosis. Hsa_circ_0005379 was significantly reduced in OSCC and showed a negative relation to tumor size and differentiation grades of OSCC. Overexpression of hsa_circ_0005379 could significantly reduce the proliferation, migration and invasion of OSCC cells, and upregulate BAX, while Bcl-2, MMP-9, and decrease cyclin D1. In addition, vimentin and N-cadherin decreased, while E-cadherin and β-catenin increased, indicating that hsa_circ_0005379 participated in the EMT process. Furthermore, the authors found that hsa_circ_0005379 may regulate the expression of epidermal growth factor receptor (EGFR) and act as an upstream molecule of the EGFR pathway. This conclusion was based on the fact that hsa_circ_0005379 can negatively regulate the expression of EGFR, but EGFR pathway agonists and inhibitors had no regulatory effect on hsa_circ_0005379.61 In another study, it was found that downregulated hsa_circ_0007059 in OSCC had similar features. Its overexpression showed a tumor-suppressive effect on OSCC and also regulated Bcl-2 family and EMT-related protein changes. But, the mechanism by which downregulated hsa_circ_0007059 promoted the growth of OSCC cells was by influencing the downstream protein levels of the AKT/mTOR signaling pathway.62 Similarly, hsa_circ_0055538 was reduced in OSCC and its overexpression could reduce the proliferation, migration and invasion of OSCC and induce OSCC cell apoptosis. After silencing hsa_circ_0055538, it was found that the levels of p53, Bax, caspase-3, etc. decreased in OSCC cells, while Bcl-2 increased. And upregulating p53 levels in OSCC cells would reverse the effect of knocking down hsa_circ_0055538 on OSCC. Therefore, the authors suggested that hsa_circ_0055538 may regulate the progression of OSCC through the p53/Bcl-2/caspase signaling pathway.63
All above, most studies have focused on detecting the expression of cell apoptosis-related proteins (BAX/Bcl-2) and EMT-related proteins (such as vimentin and N-cadherin). It suggests that circRNAs play a key role in the development of OSCC, but the specific regulatory mechanism between circRNAs and these proteins is still unclear. Some studies have revealed the protein sponge and protein scaffold capabilities of circRNAs. For instance, circMBNL1 contains multiple MBNL1 binding sites and can be specifically bound by MBNL1. Excess MBNL1 can promote circMBNL1 biogenesis, thereby reducing its mRNA level. And circMBNL1 can act as a sponge of MBNL1 to promote linear splicing of genes, thereby forming an autoregulatory loop.64 Moreover, circ-Amotl1 can act as a protein scaffold and promote PDK1-dependent phosphorylation of AKT1 by binding to PDK1 and AKT1.65 However, it has not been studied whether circRNAs in OSCC can serve as protein sponges or protein scaffolds, and further exploration will help to better understand the molecular mechanism of OSCC pathogenesis.
Therapy-Associated circRNAs
Cetuximab is an anticancer drug that specifically inhibits the cell cycle process by specifically binding to the EGFR domain of cells, thereby inducing tumor cell apoptosis. And EGFR is abundantly expressed in OSCC cells, which could promote the EMT process of OSCC, and cetuximab significantly inhibits the effect by binding to EGFR.64 Su et al found that after adding cetuximab, the early apoptosis rates of OSCC cells with overexpression of hsa_circ_0005379 increased to 2-fold compared with the control group, indicating that overexpression of hsa_circ_0005379 could increase the sensitivity of cetuximab to OSCC.61 Fucoidan is a sulfated polysaccharide isolated from various brown algae and brown algae, and has many biological effects including anti-cancer effects.66,67 Zhang et al found that fucoidan could inhibit OSCC growth, migration and invasion.68 In addition, bioinformatics database prediction and qPCR experiments confirmed that fucoidan can increase the expression of circFLNA in OSCC cell lines. Overexpression of circFLNA also inhibited the proliferation, migration and invasion of OSCC and induced apoptosis. After silencing circFLNA in OSCC cells, the antitumor effect of fucoidan was reversed, suggesting that the fucoidan-circFLNA axis may regulate the process of OSCC. Chen et al identified that circATRNL1 was significantly downregulated in OSCC and its overexpression could increase the radiosensitivity of OSCC cells by inhibiting cell survival and inducing apoptosis.69 Further experiments revealed that circATRNL1 was a sponge of miR-23a-3p, which can enhance the expression of its downstream target PTEN. In addition, knocking down PTEN increased the colony-forming ability and proliferation of OSCC cells, and inhibited cell viability, apoptosis as well as cell-cycle arrest, while circATRNL1 overexpression caused the opposite effect. Therefore, they believed that circATRNL1/miR-23a-3p/PTEN axis may play an important role in regulating the radiosensitivity of OSCC. However, it is not clear whether all of the above circRNAs are beneficial to clinical treatment, and a large number of experiments are still required to verify their effects before entering the clinic. Especially when used in humans, safety research is particularly important, and current research still lacks relevant experiments.
Conclusions and Perspectives
Extensive exploration in the field of circRNA research has brought us many new insights into cancer pathobiology. Advanced sequencing technologies can also help researchers better explore circRNAs and predict their functions, thus greatly promoting the development of this field. In addition, with the maturation of experimental verification, the functional research of circRNAs on cancer biological behavior has been greatly promoted. Mounting evidence have revealed the important role of circRNAs in the development of OSCC. They have great potential as biomarkers and may be used to diagnose and predict the prognosis of OSCC patients. And circRNAs can act as miRNA sponges to regulate the expression of downstream oncogenic molecules via circRNAs-miRNAs-mRNAs axis, suggesting that circRNAs play a key role in regulating the progression of OSCC. CircRNAs can also affect cancer-related signaling pathways in OSCC, such as MAPK and PI3K/Akt/mTOR axis, and might have an impact on the chemosensitivity and radiosensitivity of OSCC.
However, the study of circRNAs in OSCC is still in its infancy. First, circRNAs, like other ncRNAs as potential biomarkers, have more complicated design and identification methods in experiments, and these technologies need to be standardized. Second, there is still a lack of clinical applications on circRNAs, and the current research conclusions are only based on the results of a few OSCC patients. Hence, their accuracy and replicability as biomarkers should be validated, and this is crucial for their clinical application in the future. Importantly, current circRNAs exhibit different correlations with the early or advanced stages of OSCC. The underlying differences are interesting and seem worth of further exploration. Third, most existing research on circRNAs in OSCC focuses on circRNAs located in the cytoplasm and their function as miRNA sponges. Nevertheless, there is currently no research on their protein sponges or protein scaffolds capabilities, and these functions have been widely explored in other fields.70–72 Fourth, circRNA can also be a template for translation, and Chen et al have developed the first protein-coding circRNA database.73 Circ_ FBXW774 can encode a novel protein, which is named FBXW7-185aa, and can inhibit the proliferation and cell cycle of glioma cells. CircRNA_FECR175 located in the nucleus could control the growth of breast cancer by regulating DNA methylase and demethylase. After screening out the most differentially expressed circRNAs, through comprehensive analyzing the characteristic sequences they contain (such as MREs, RBPs motifs, ORF and IRES) and the analysis of existing databases, the function of circRNAs can be well predicted. However, more should be done in OSCC in the future.
In addition, circRNAs can be released through extracellular vesicles and may play a role in intercellular communication and TME.76 Hsa_circRNA_002178 is highly expressed in lung adenocarcinoma and can be detected in plasma exosomes, showing potential as a diagnostic biomarker.77 But the study of circRNAs in extracellular vesicles is still blank in OSCC, and future research in this field will be of great significance to the diagnosis and pathogenesis of OSCC. Interestingly, artificial circRNAs have also been found to function in eukaryotic cells. For example, engineered circRNA is designed to treat cardiac hypertrophy by sponging to target miRNA, and shows exciting potential as future therapeutics.78 Besides, it can enhance PDL1 expression through sponging miR-34 and may be used as a target for immunotherapy. And increased evidence has uncovered the vital role of circRNAs in the sensitivity of radiotherapy and chemotherapy.79 In conclusion, the results of these studies on circRNAs are inspiring, and fully clarifying the role of circRNAs will hope to accelerate circRNAs from bench to bed, especially in OSCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. van Zyl A, Bunn BK. Clinical features of oral cancer. SADJ. 2012;67(10):566–569.
2. Mehrotra R, Yadav S. Oral squamous cell carcinoma: etiology, pathogenesis and prognostic value of genomic alterations. Indian J Cancer. 2006;43(2):60–66. doi:10.4103/0019-509X.25886
3. WHO Cancer Key Facts. Available from: https://www.who.int/news-room/fact-sheets/detail/oral-health. 2018.
4. Nor JE, Gutkind JS. Head and neck cancer in the new era of precision medicine. J Dent Res. 2018;97(6):601–602. doi:10.1177/0022034518772278
5. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
6. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
7. Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):8. doi:10.1186/s12943-019-1113-0
8. Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50. doi:10.1016/j.canlet.2018.01.011
9. Zhang Q, Wang W, Zhou Q, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi:10.1186/s12943-019-1125-9
10. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. doi:10.1186/s12943-020-1135-7
11. Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
12. Bach D-H, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
13. Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol. 2018;9:1262. doi:10.3389/fphys.2018.01262
14. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16(8):503–514. doi:10.1038/s41569-019-0185-2
15. Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi:10.1016/j.molcel.2015.03.027
16. Dong R, Ma X-K, Chen L-L, Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14(8):1064–1074. doi:10.1080/15476286.2016.1269999
17. Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi:10.7150/ijms.28047
18. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
19. Zhang H-D, Jiang L-H, Sun D-W, Hou J-C, Ji Z-L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
20. Hsiao K-Y, Sun HS, Tsai S-J. Circular RNA – new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141. doi:10.1177/1535370217708978
21. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. doi:10.7150/jca.30828
22. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi:10.1093/nar/gkw027
23. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi:10.1038/s41388-017-0019-9
24. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71. doi:10.1186/s12943-019-0969-3
25. Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. doi:10.1038/s41467-018-06862-2
26. Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47. doi:10.1186/s12943-019-1010-6
27. Zhang L, Meng X, Zhu X-W, et al. Long non-coding RNAs in oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol Cancer. 2019;18(1):102.
28. D’Souza W, Kumar A. microRNAs in oral cancer: moving from bench to bed as next generation medicine. Oral Oncol. 2020;111:104916. doi:10.1016/j.oraloncology.2020.104916
29. Verduci L, Ferraiuolo M, Sacconi A, et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18.
30. Wang Y-F, Li B-W, Sun S, et al. Circular RNA expression in oral squamous cell carcinoma. Front Oncol. 2018;8:398. doi:10.3389/fonc.2018.00398
31. Deng W, Peng W, Wang T, et al. Microarray profile of circular RNAs identifies hsa_circRNA_102459 and hsa_circRNA_043621 as important regulators in oral squamous cell carcinoma. Oncol Rep. 2019;42(6):2738–2749. doi:10.3892/or.2019.7369
32. Qiu X, Ke X, Ma H, et al. Profiling and bioinformatics analyses reveal differential expression of circular RNA in tongue cancer revealed by high-throughput sequencing. J Cell Biochem. 2019;120(3):4102–4112. doi:10.1002/jcb.27695
33. Zhao S-Y, Wang J, Ouyang S-B, Huang Z-K, Liao L. Salivary circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell Physiol Biochem. 2018;47(6):2511–2521. doi:10.1159/000491624
34. Ganci F, Sacconi A, Manciocco V, et al. MicroRNA expression as predictor of local recurrence risk in oral squamous cell carcinoma. Head Neck. 2016;38(Suppl 1):E189–E197. doi:10.1002/hed.23969
35. Li B, Wang F, Li X, Sun S, Shen Y, Yang H. Hsa_circ_0008309 may be a potential biomarker for oral squamous cell carcinoma. Dis Markers. 2018;2018:7496890. doi:10.1155/2018/7496890
36. Wang Z, Tang J, Wang Y, et al. Circular RNA hsa_circ_009755 downregulation correlates with clinicopathology in oral squamous cell carcinoma. Onco Targets Ther. 2019;12:4025–4031. doi:10.2147/OTT.S196618
37. Sun S, Li B, Wang Y, et al. Clinical significance of the decreased expression of hsa_circ_001242 in oral squamous cell carcinoma. Dis Markers. 2018;2018:6514795. doi:10.1155/2018/6514795
38. Dou Z, Li S, Ren W, et al. Decreased expression of hsa_circ_0072387 as a valuable predictor for oral squamous cell carcinoma. Oral Dis. 2019;25(5):1302–1308. doi:10.1111/odi.13094
39. Ouyang S-B, Wang J, Zhao S-Y, Zhang X-H, Liao L. CircRNA_0109291 regulates cell growth and migration in oral squamous cell carcinoma and its clinical significance. Iran J Basic Med Sci. 2018;21(11):1186–1191. doi:10.22038/IJBMS.2018.30347.7313
40. Li L, Zhang Z-T. Hsa_circ_0086414 might be a diagnostic biomarker of oral squamous cell carcinoma. Med Sci Monitor. 2020;26.
41. Gao L, Wang QB, Zhi Y, et al. Down-regulation of hsa_circ_0092125 is related to the occurrence and development of oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2019;S0901-5027(0919):31262–31265.
42. Peng Q-S, Cheng Y-N, Zhang W-B, Fan H, Mao Q-H, Xu P. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway. Cell Death Dis. 2020;11(2):112. doi:10.1038/s41419-020-2273-y
43. Shao B, He L. Hsa_circ_0001742 promotes tongue squamous cell carcinoma progression via modulating miR-634 expression. Biochem Biophys Res Commun. 2019;513(1):135–140. doi:10.1016/j.bbrc.2019.03.122
44. Li X, Zhang H, Wang Y, Sun S, Shen Y, Yang H. Silencing circular RNA hsa_circ_0004491 promotes metastasis of oral squamous cell carcinoma. Life Sci. 2019;239:116883. doi:10.1016/j.lfs.2019.116883
45. He T, Li X, Xie D, Tian L. Overexpressed circPVT1 in oral squamous cell carcinoma promotes proliferation by serving as a miRNA sponge. Mol Med Rep. 2019;20(4):3509–3518. doi:10.3892/mmr.2019.10615
46. Gao L, Zhao C, Li S, et al. circ-PKD2 inhibits carcinogenesis via the miR-204-3p/APC2 axis in oral squamous cell carcinoma. Mol Carcinog. 2019;58(10):1783–1794. doi:10.1002/mc.23065
47. Wang D, Berglund A, Kenchappa RS, Forsyth PA, Mulé JJ, Etame AB. BIRC3 is a novel driver of therapeutic resistance in Glioblastoma. Sci Rep. 2016;6:21710. doi:10.1038/srep21710
48. Wang L, Wei Y, Yan Y, et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39(3):951–966.
49. Zhu X, Shao P, Tang Y, Shu M, Hu W-W, Zhang Y. hsa_circRNA_100533 regulates GNAS by sponging hsa_miR_933 to prevent oral squamous cell carcinoma. J Cell Biochem. 2019;120(11):19159–19171.
50. Hollstein PE, Shaw RJ. GNAS shifts metabolism in pancreatic cancer. Nat Cell Biol. 2018;20(7):740–741. doi:10.1038/s41556-018-0120-5
51. Lee H, La B-M, Hwang I, Kang Y-N, Choi I-J, Lee J-H. Absence of GNAS mutation in colorectal carcinogenesis. Tumori. 2017;103(2):209–211. doi:10.5301/tj.5000400
52. Xia B, Hong T, He X, Hu X, Gao Y. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplant. 2019;28(12):1614–1623. doi:10.1177/0963689719875409
53. Zhao W, Cui Y, Liu L, et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2019. doi:10.1038/s41418-41019-40423-41415
54. Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol. 2019;234(11):19130–19140. doi:10.1002/jcp.28692
55. Wang Y, Zhang X, Wang Z, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi:10.1016/j.canlet.2018.07.016
56. Kang -Y-Y, Sun F-L, Zhang Y, Wang Z. SIRT1 acts as a potential tumor suppressor in oral squamous cell carcinoma. J Chin Med Assoc. 2018;81(5):416–422. doi:10.1016/j.jcma.2017.09.004
57. Wang Y, Ma Y, Zhu A, Zhao H, Liao L. Accurate facade feature extraction method for buildings from three-dimensional point cloud data considering structural information. ISPRS J Photogrammetry Remote Sens. 2018;139:146–153. doi:10.1016/j.isprsjprs.2017.11.015
58. Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–2092. doi:10.2147/OTT.S233672
59. Zheng XB, Zhang M, Xu MQ. Detection and characterization of ciRS-7: a potential promoter of the development of cancer. Neoplasma. 2017;64(3):321–328. doi:10.4149/neo_2017_301
60. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi:10.1158/0008-5472.CAN-13-1568
61. Su W, Wang Y, Wang F, et al. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer. 2019;19(1):400. doi:10.1186/s12885-019-5593-5
62. Su W, Wang Y, Wang F, et al. Circular RNA hsa_circ_0007059 indicates prognosis and influences malignant behavior via AKT/mTOR in oral squamous cell carcinoma. J Cell Physiol. 2019;234(9):15156–15166. doi:10.1002/jcp.28156
63. Su W, Sun S, Wang F, Shen Y, Yang H. Circular RNA hsa_circ_0055538 regulates the malignant biological behavior of oral squamous cell carcinoma through the p53/Bcl-2/caspase signaling pathway. J Transl Med. 2019;17(1):76. doi:10.1186/s12967-019-1830-6
64. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
65. Zeng Y, Du WW, Wu Y, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7(16):3842–3855. doi:10.7150/thno.19764
66. Li B, Lu F, Wei X, Zhao R. Fucoidan: structure and bioactivity. Molecules. 2008;13(8):1671–1695. doi:10.3390/molecules13081671
67. van Weelden G, Bobiński M, Okła K, van Weelden WJ, Romano A, Pijnenborg JMA. Fucoidan structure and activity in relation to anti-cancer mechanisms. Mar Drugs. 2019;17(1):32. doi:10.3390/md17010032
68. Zhang N, Gao L, Ren W, et al. Fucoidan affects oral squamous cell carcinoma cell functions in vitro by regulating FLNA-derived circular RNA. Ann N Y Acad Sci. 2019. doi:10.1111/nyas.14190
69. Chen G, Li Y, He Y, et al. Upregulation of Circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol Ther Nucleic Acids. 2020;19:961–973. doi:10.1016/j.omtn.2019.12.031
70. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. doi:10.1002/jnr.24356
71. Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi:10.1038/cdd.2016.133
72. Xia P, Wang S, Ye B, et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48(4):688–701.e687. doi:10.1016/j.immuni.2018.03.016
73. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi:10.1038/srep34985
74. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–315. doi:10.1093/jnci/djx166
75. Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218. doi:10.1186/s13059-018-1594-y
76. Preußer C, Hung LH, Schneider T, et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7(1):1424473. doi:10.1080/20013078.2018.1424473
77. Wang J, Zhao X, Wang Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. doi:10.1038/s41419-020-2230-9
78. Lavenniah A, Luu TDA, Li YP, et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol Ther. 2020;28(6):1506–1517. doi:10.1016/j.ymthe.2020.04.006
79. Cui C, Yang J, Li X, Liu D, Fu L, Wang X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19(1):58. doi:10.1186/s12943-020-01180-y
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.