Back to Journals » OncoTargets and Therapy » Volume 10
CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway
Authors Chen G, Shi Y, Zhang Y, Sun J
Received 2 September 2017
Accepted for publication 2 November 2017
Published 5 December 2017 Volume 2017:10 Pages 5783—5794
DOI https://doi.org/10.2147/OTT.S150678
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Genwen Chen,1,* Yanting Shi,2,* Yu Zhang,1 Jianyong Sun1
1Department of Gastroenterology, Zhongshan Hospital, Fudan University, Shanghai, 2Department of Nephrology, Zhongshan Hospital, Fudan University, Shanghai, China
*These authors contributed equally to this work
Abstract: Circular RNAs (circRNAs) are a novel class of noncoding RNAs that play an important role in cancer. However, the mechanisms by which circRNAs regulate gene expression in pancreatic ductal adenocarcinoma (PDAC) remain unclear. This study seeks to elucidate the role that circRNAs play in the proliferation of PDAC cells. On the basis of previous studies of circRNA expression profiles in PDAC, we found that the circRNA_100782 was markedly upregulated in PDAC tissue. Functional experiments revealed that circRNA_100782 downregulation inhibited BxPC3 cell proliferation and colony formation. Loss-of-function studies showed that knockdown of circRNA_100782 inhibited cell proliferation by downregulating the microRNA-124 (miR-124) target genes interleukin-6 receptor (IL6R) and signal transducer and activator of transcription 3 (STAT3). Overexpression of miR-124 also inhibited BxPC3 cell proliferation by reducing the expression of IL6R and STAT3, which was consistent with the result of silencing circRNA_100782. In addition, luciferase assay revealed that miR-124 was a direct target of circRNA_100782. Silencing STAT3 inhibited BxPC3 cell proliferation and colony formation. Cell viability was reduced in BxPC3 cells treated with si-circRNA_100782 and miR-124 mimic, and this effect could be attenuated by activating STAT3. In vivo study validated that circRNA_100782 knockdown suppressed BxPC3 xenografts in nude mice. Taken together, these results suggest that circRNA_100782 regulates BxPC3 cell proliferation by acting as miR-124 sponge through the IL6–STAT3 pathway.
Keywords: circular RNA, pancreatic ductal adenocarcinoma, sponge, proliferation, microRNA, IL6-STAT3 pathway
Introduction
Circular RNAs (circRNAs) are a novel class of noncoding RNAs that are formed by a junction of the 5′end and 3′end.1 CircRNAs were discovered a few decades ago, but they were neither paid enough attention nor investigated until recently.2 CircRNA, as well as microRNA (miRNA) and long noncoding RNA are involved in the process of transcriptional and post-translational gene expression. Mounting evidence has shown that circRNAs are dysfunctional in nervous system disease,3 heart disease4 and human cancer.5 There is increasing evidence showing that circRNAs are involved in tumorigenesis, proliferation, apoptosis, migration, invasion and metastasis in human carcinomas.6–8 Additionally, circRNAs are enriched and stable in many tissues and exosomes, suggesting that they are promising biomarkers for cancer diagnosis and prognosis.7,9,10 However, the role of circRNAs in pancreatic ductal adenocarcinoma (PDAC) remains unclear.
miRNAs are a type of noncoding RNA of ~20 nucleotides and negatively regulate multiple genes by interacting with 3′-untranslated regions (3′-UTR) of cognate mRNA targets that play crucial roles in post-transcriptional regulation.11 In the past, miRNAs have been widely investigated and identified as key regulators and biomarkers in cancers. miRNAs act as oncogenes or tumor suppressors by altering the expression of their targets and signaling pathways both in vivo and in vitro.12,13 Recent research has revealed that circRNAs can act as miRNA sponges and regulate the targets of miRNAs.14 However, to our knowledge, there are no previous reports concerning circRNA–miRNA interaction in PDAC.
PDAC ranks as the third leading cause of cancer death in the USA. Due to the lack of early diagnosis and effective therapies, PDAC has one of the worst prognoses.15,16 Signal transducer and activator of transcription 3 (STAT3) acts as an oncogene in multiple tumors.17 Notably, activation of STAT3 leads to malignancy of tumorigenesis, cell proliferation and migration.18,19 The interleukin-6 receptor (IL6R) engages through gp 130 ligands and activates the janus kinase 2 (JAK2) family. Phosphorylation of JAK2 (p-JAK2) then activates the STAT3 proteins.20 Previous studies have revealed that the IL6-JAK2-STAT3 signaling pathway is crucial for cell growth and survival.21,22 This pathway was frequently activated in pancreatic cell lines and PDAC tissue specimens.20 Lesina et al showed that the IL6-JAK2-STAT3 signaling pathway improved proliferation and oncogenesis in PDAC both in vivo and in vitro.23 IL6R is upstream of STAT3, and upregulation of IL6R activates the phosphorylation of STAT3 (p-STAT3).24
This study focused on a microarray analysis of the expression of circular RNA-has_circRNA_100782, which was upregulated in PDAC tissues (GEO No: 69362: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69362). CircRNA_100782 is encoded by the sequence located at chr11:33307958–33309057 and has a spliced length of 1,099 nt. CircRNA_100782 was first found upregulated in PDAC and in the PDAC cell line BxPC3 instead of human pancreatic duct epithelial cells (HPDE). Our research sought to investigate the role of circRNA_100782 in PDAC and explore the mechanism of circRNA_100782 in cell proliferation from 3 aspects of “circRNA-miRNA-target gene”.
Materials and methods
Cell lines and cell culture
The PDAC cell lines BxPC3 and HPDE were obtained from American Type Culture Collection (Manassas, VA, USA). HEK293T cells were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). BxPC3 and HPDE cells were cultured in Roswell Park Memorial Institute 1640 (HyClone Laboratories, Logan, UT, USA) and HEK293T were cultured in DMEM (HyClone Laboratories) with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), streptomycin (100 μg/mL) and penicillin (100 μg/mL) (Gibco). Cells were grown in a humidified 5% CO2 incubator at 37°C.
Vector construction
For the luciferase assay, the coding sequence of circRNA_100782 was synthesized and inserted into the NheI and SalI sites directly downstream of a cytomegalovirus promoter-driven firefly luciferase cassette in the pmirGLO vector (Promega, Madison, WI, USA). The recombinant luciferase reporter plasmids contained full-length sequences of circRNA_100782. The empty pmirGLO vector was used for negative control. The constructs were confirmed by DNA sequencing (Sangon Biotech, Shanghai, China).
Cell proliferation assay and colony formation assay
The Cell Counting Kit-8 (CCK-8, Dojindo Chemical Laboratory, Kumamoto, Japan) and colony formation assay were conducted to determine the cell proliferation activity. Approximately 5×103 cells per well were transferred to 96-well plates after transfection and were subjected to the CCK-8 assay. After incubation at 37°C for 1.5 h, the absorbance at 450 nm was measured following the addition of 10 μL of the CCK-8 solution. There were 5 replicates for each group, and 3 independent experiments were performed. For the colony formation assay, 500–1,000 cells were seeded in 6-well plates overnight. The cells were transfected with siRNA or miR-124 mimic for 24 h. The medium was removed and replaced with fresh medium. When the colonies were visible without a microscope, cell colonies were fixed with 4% paraformaldehyde and stained using 0.1% crystal violet. The number of colonies was quantified by counting the colonies with more than 50 cells.
Cell transfection
INTERFERin (Polyplus transfection, Illkirch, France) was used for the transfection experiments with miRNA mimics or siRNAs according to the manufacturer’s instructions. miR-124 mimic, miR-124 inhibitor, negative control miRNA and siRNAs targeting circRNA_100782, STAT3 and siRNA negative control were obtained from RiboBio (Guangzhou, China). The knockdown efficiency of circRNA_100782 was evaluated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Western blot was used to measure the efficiency of si-STAT3. To identify the role of IL6 in si-circRNA_100782 and miR-124-induced cell suppression, BxPC3 cells were treated with recombinant human IL6 cytokine (PeproTech, Rocky Hill, NJ, USA) after transfection with si-circRNA_100782 and miR-124 according to the manufacturer’s instructions.
RNA isolation and real-time transcription PCR
Total RNA was isolated using TRIzol regent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. For circRNA_100782, IL6R and STAT3 detection, cDNA was synthesized using the Prime Script RT reagent Kit (Takara Bio, Shiga, Japan). qRT-PCR was performed in a 96-well plate format using SYBR green dye (Takara Bio, Dalian, China) by ABI Prism 7500 Fast Real-time PCR System (Applied Biosystems, Foster City, CA, USA) following the procedure according to the manufacturer’s instructions. For miR-124 detection, reverse transcription of 1 μg RNA was performed using miRNA reverse transcription kit with miR-124-specific stem-loop primer (RiboBio). qRT-PCR amplification was performed using the Bulge-Loop miRNA qRT-PCR Starter Kit (RiboBio). The levels of circRNA_100782 and mRNAs were normalized by ACTB, while miR-124 was normalized against U6 and calculated using the 2−ΔΔCt method. The primers used are listed below: 5′-TATGTTGGTGGATCCTGTTCGGCA-3′(forward), 5′-TGGTGGGTAGACCAAGACTTGTGA-3′(reverse) for circRNA_100782, yielding a product of 146 bp. 5′-CATGTACGTTGCTATCCAGGC-3′(forward), 5′-CTCCTTAATGTCACGCACGAT-3′(reverse) for ACTB. 5′-CAGCAGCTTGACACACGGTA-3′(forward), 5′-AAACACCAAAGTGGCATGTGA-3′(reverse) for STAT3. 5′-TACACCAACCTCTCGTACATCG-3′(forward), 5′-CATGTCTGAAGCGCAGTAAGATT-3′(reverse) for IL6R.
Western blot
Total protein from frozen tissues and cells was extracted using radio-immunoprecipitation assay lysis buffer, and the lysates were cleared by centrifugation (12,000× g, 4°C, 15 min). The bicinchoninic acid protein assay (Beyotime Biotechnology, Shanghai, China) was used to measure the total protein level. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfected to a polyvinylidene fluoride membrane (Immobion-P Transfer Membrane, Millipore Corp., Billerica, MA, USA). After blocking the membrane with 5% non-fat dry milk for 1 h, the membrane was incubated overnight at 4°C with antibodies against STAT3, p-STAT3 (Y705), JAK2, p-JAK2, glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1:1,000, Cell Signaling Technology, Danvers, MA, USA) and IL6R (1:500 RD Systems, Minneapolis, MN, USA). After washing with tris-buffered saline with Tween-20, specific secondary anti-rabbit antibodies or anti-mouse IgG secondary antibodies conjugated to horseradish peroxidase (Cell Signaling Technology) were incubated at room temperature for 1 h. Protein bands were visualized using the enhanced chemiluminescence ECL method and analyzed with Quantity One 4.6.2 software (Bio-Rad, Hercules, CA, USA).
Luciferase reporter assay
HEK293T cells were cotransfected with 500 ng of pmirGLO-circRNA_100782 or pmirGLO-vector with 50 nm of miR-124 mimic in 24-well plates. The combined plasmids and miR-124 mimic were transfected into HEK293T cells using Lipofectamine 3000 (Thermo Scientific). After 48 h of incubation, cells were collected and the firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol.
Lentiviral vector production and cell infection
Lentivirus carrying hairpin-silencing RNA to circRNA_100782 (sh-circRNA_100782) and sh-circRNA_100782 control (sh-Ctrl) was packaged in HEK293T cells and the supernatant was concentrated. For stable knockdown of circRNA_100782, BxPC3 cells were infected with virus supernatant with 8 μg/mL of polybrene. After 24 h infection, the medium was replaced with fresh medium containing 2 μg/mL of puromycin.
Tumor formation in nude mice
Ten 6-week-old male BALB/c nude mice were divided into 2 groups randomly. Each group was composed of 5 mice that were injected with 2×106 control cells (sh-Ctrl) or circRNA_100782 knockdown cells (sh-circRNA_100782). The tumor volume was calculated by measuring the length and width of the tumor (tumor volume =1/2 length × width2) every week. After 5 weeks, the mice were sacrificed by anesthesia and the tumors were removed and weighed for further analysis. All animal studies were approved by the Animal Ethics Committee of Zhongshan Hospital, Fudan University and experiments were conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (US National Research Council Publication 85–23, revised 2011).
Statistical analysis
Data from at least 3 separate experiments are expressed as mean ± SD. The results were analyzed using the Student’s t-test and one-way analysis of variance. Statistical analyses were performed using software SPSS version 18.0 (SPSS, Chicago, IL, USA), and p<0.05 was considered statistically significant.
Results
CircRNA_100782 is upregulated in BxPC3 compared with HPDE cell line
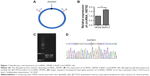
We first investigated circRNA_100782 expression between the normal pancreatic epithelial cell line (HPDE) and BxPC3 by qRT-PCR using divergent primer (Figure 1A). We found that the circRNA_100782 level was higher in BxPC3 than HPDE cells (Figure 1B). To confirm the product of qRT-PCR, we performed agarose gel electrophoresis and a single band of 146 bp was observed (Figure 1C). Additionally, the back-splice junction of circRNA_100782 was confirmed by Sanger sequence (Figure 1D), which was consistent with the sequence from circbase.25
The function of circRNA_100782 in BxPC3 cell line
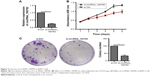
Knowing that circRNA_100782 is upregulated in PDAC tissue and BxPC3 cell line, we wanted to explore the effect of circRNA_100782 in vitro. We silenced circRNA_100782 using siRNA that targeted the back-splice sequence. The expression of circRNA_100782 decreased significantly as expected (Figure 2A). CCK-8 assay showed that knockdown of circRNA_100782 decreased BxPC3 cell proliferation (Figure 2B). The colony formation assay revealed that knockdown of circRNA_100782 significantly inhibited cell colony formation. The colony formation numbers on the plates were found to be decreased in BxPC3 cells transfected with si-circRNA_100782 compared with the control (Figure 2C). This indicates that circRNA_100782 is a cell proliferation regulator in PDAC.
CircRNA_100782 functions by binding to miR-124
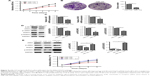
CircRNA can bind to miRNA and then regulate miRNA target genes by serving as miRNA sponges.1,26,27 We then wanted to determine whether circRNA_100782 could act as miRNA sponge. We screened miRNAs that may have binding sites targeting circRNA_100782 sequence based on TargetScan (www.targetscan.org) database and found that miR-124 has a binding site with circRNA_100782 (Figure 3A). miR-124 has been proven as an anti-oncogenesis miRNA in many cancers, including hepatocellular carcinoma (HCC),28 esophageal cancer,29 PDAC,30 and we found that miR-124 was downregulated in BxPC3 cells compared with HPDE cells (Figure 3B), which suggested the antigenic role of miR-124 in BxPC3. We then investigated whether circRNA_100782 acts as miR-124 sponge and inhibits its function. To confirm whether circRNA_100782 can bind miR-124, a dual-luciferase reporter system was performed. Luciferase-circRNA_100782 reporter was cotransfected with miR-124 mimic in HEK293T cells that resulted in reduction of luciferase activity by 30% compared with vector-Ctrl (Figure 3C and D).
Based on the relationship between circRNA_100782 and miR-124, we explored whether circRNA_100782 could regulate miR-124 target genes. Previous studies found that miR-124 acts as a tumor suppressor gene in various cancer types. Searching target prediction software (TargetScan), we found that IL6R and STAT3 are predictive targets of miR-124. Previous studies have demonstrated that miR-124 functions by targeting 3′-UTR of IL6R and STAT3.11,24 IL6R is the upstream of p-STAT3 and the IL6-STAT3 pathway plays a crucial role in PDAC. Then we hypothesized that circRNA_100782 may function by interacting with miR-124 and then activating the IL6-JAK2-STAT3 signaling pathway. Next, we investigated the correlation between circRNA_100782 and the miR-124 target genes IL6R and STAT3. Knockdown of circRNA_100782 decreased the mRNA levels of IL6R and STAT3 (Figure 3E). Western blot confirmed a decrease in protein levels for both IL6R and STAT3. The levels of p-STAT3 and p-JAK2 also decreased significantly, which was induced by reduction in IL6R (Figure 3F). Downregulation of IL6R, STAT3 and p-STAT3 by si-circRNA_100782 could be abrogated by miR-124 inhibitor (Figure 3G). Interestingly, qRT-PCR showed that circRNA_100782 knockdown had no obvious effect on miR-124, and ectopic expression of miR-124 did not decrease the level of circRNA_100782 (data not shown). This was in accordance with the ciRS-7 (circular RNA sponge for miR-7) findings.1 These data indicate that circRNA_100782 may activate IL6R and STAT3 by binding to miR-124 and inhibiting miR-124 activity.
Next, we investigated whether the IL6-JAK2-STAT3 signaling pathway was responsible for BxPC3 cell proliferation. Western blot confirmed that STAT3 is activated in BxPC3 cells than in HPDE cells (Figure 4A). Ectopic expression of IL6 significantly activated p-JAK2 and p-STAT3 and increased BxPC3 cell proliferation (Figure 4B and C). Furthermore, knockdown of STAT3 decreased BxPC3 cell proliferation and colony formation (Figure 4D–F). To evaluate whether IL6-JAK2-STAT3 pathway was responsible for circRNA_100782-induced cell proliferation, we activated STAT3 by ectopic expression of IL6, which significantly rescued the protein levels of p-JAK2 and p-STAT3 under repression by si-circRNA_100782 (Figure 4G). In addition, IL6 abolished the anti-proliferation effect of si-circRNA_100782 (Figure 4H). Taken together, circRNA_100782 may activate IL6R and STAT3 by binding to miR-124 and may inhibit miR-124 activity.
MiR-124 mediates BxPC3 cell proliferation through the IL6-JAK2-STAT3 pathway
Although miR-124 has been demonstrated to be an anti-oncogenesis miRNA, its role on BxPC3 cells remains unknown. To investigate whether miR-124 is responsible for the downregulation of IL6R and STAT3, we transfected miR-124 mimic into BxPC3 cells. Similar to the experiment performed with si-circRNA_100782, overexpression of miR-124 significantly repressed BxPC3 cell proliferation and colony formation (Figure 5A and B). Likewise, miR-124 reduced both the mRNA levels of IL6R and STAT3 and the protein levels of IL6R, p-JAK2, STAT3 and p-STAT3 (Figure 5C and D). As expected, IL6 abolished the miR-124-induced reduction of cell viability and p-JAK2 and p-STAT3 levels (Figure 5E and F). Taken together, these results indicate that circRNA_100782 regulates the IL6-JAK2-STAT3 pathway by acting as miR-124 sponge.
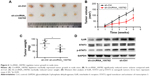
CircRNA_100782 knockdown repressed BxPC3 xenograft growth in vivo
We further investigated the biological function of circRNA_100782 in BxPC3 cell growth in BALB/c nude mice (5 mice per group). Each nude mouse was injected with 2×106 of sh-Ctrl BxPC3 cells or sh-circRNA_100782 BxPC3 cells. After 5 weeks, the mice were sacrificed and we found that circRNA_100782 knockdown repressed tumor growth significantly. The sh-Ctrl cells grew faster than sh-circRNA_100782 group (Figure 6A–C). To determine whether circRNA_100782 functions through STAT3 pathway, Western blot was performed to measure the expression levels of miR-124 targets. We found that the expression of IL6R, STAT3 and p-STAT3 was downregulated in the sh-circRNA_100782 BxPC3 xenografts (Figure 6D). These data indicate that circRNA_100782 knockdown repressed BxPC3 cell growth in vivo through the IL6-STAT3 pathway.
Discussion
In this study, we identified circRNA_100782 to be a molecule that promotes BxPC3 pancreatic cancer cell proliferation by targeting miR-124 and its target genes IL6R and STAT3. We found that miR-124 was a negative regulator of proliferation in BxPC3 cells. Activating STAT3 attenuates the anti-proliferation induced by si-circRNA_100782 or miR-124. Our data from the “circRNA-miRNA-target gene” perspective demonstrate that circRNA_100782 regulates BxPC3 cell proliferation by interacting with miR-124 through the IL6-STAT3 pathway.
Due to their special structure, circRNAs can act as promising noncoding RNAs to regulate gene expression. The classic role of circRNAs is to serve as miRNA sponge, as in the case of circRNA sex-determining region Y (Sry) and ciRS-7.14 They bind to miRNAs and act as candidates for competing endogenous RNAs. ciRS-7 harbors 63 miR-7 seed matches and acts as post-transcriptional regulator in brain tissue. However, ciRS-7 does not affect the miR-7 expression level.1 This is consistent with our study; knockdown of circRNA_100782 does not increase the expression of miR-124; instead, it decreases the expression of miR-124 targets. Circular RNA MTO1 (circMTO1) acts as a sponge for miR-9, suppressing HCC progression, and reduced circMTO1 expression predicts a decreased survival time. Silencing circMTO1 could downregulate the miR-9 target gene p21, which results in the promotion of HCC cell proliferation and invasion.7 In CRC, has_circ_001569 sponged with miR-145 and promoted miR-145 target genes E2F5, BAG4 and FMNL2 then acted as a positive regulator in cell proliferation and invasion.31 circPVT1 acts as a proliferative factor and a good prognostic marker in gastric cancer.32 This indicates that circRNA can act as a potential diagnostic and therapeutic target for tumors.
Based on the circRNA expression profile, we found that circRNAs are significantly dysregulated in PDAC. Since circRNA_100782 was obviously upregulated, we proposed that circRNA_100782 may have a functional role in PDAC. We found that circRNA_100782 was more upregulated in BxPC3 than in HPDE cells, and circRNA_100782 knockdown significantly decreased BxPC3 proliferation. Since circRNA functions by binding with miRNA, we wanted to determine whether circRNA_100782 could serve as miRNA sponge. Bioinformatic analysis predicted that circRNA_100782 had a binding site with miR-124. Additionally, it has been proven that miR-124 is a tumor suppressor in many cancers. Therefore, we carried out luciferase assay and found that cotransfecting with miR-124 could reduce luciferase activity. qRT-PCR and Western blot confirmed that silencing circRNA_100782 decreased the levels of IL6R, p-JAK2, STAT3 and p-STAT3. The downregulation of IL6R, p-JAK2, STAT3 and p-STAT3 could be rescued by miR-124 inhibitor. We further investigated the function of miR-124 in BxPC3 cell line. Overexpression of miR-124 greatly inhibited BxPC3 cell proliferation and colony formation. Consistent with previous studies, miR-124 directly targets the 3′-UTR of IL6R and STAT3.11 The IL6R, p-JAK2, STAT3, and p-STAT3 protein levels were significantly downregulated by the transient overexpression of miR-124. In addition, miR-124 target genes positively correlated with circRNA_100782. IL6 activation of STAT3 rescued cell proliferation and the protein levels of p-JAK2 and p-STAT3 reduced by si-circRNA_100782 and miR-124. In vivo study confirmed the role of circRNA_100782 in vitro. Tumors were significantly suppressed by knockdown of circRNA_100782. Taken together, these data suggest that circRNA_100782 regulates BxPC3 cell proliferation by acting as miR-124 sponge through IL6R and STAT3.
Conclusion
Our study reveals that circRNA_100782 plays a crucial role in PDAC. CircRNA_100782 knockdown suppresses BxPC3 cell proliferation. CircRNA_100782 interacts with miR-124 and regulates the IL6-STAT3 pathway. The IL6-STAT3 pathway is responsible for circRNA_100782 and miR-124-mediated repression of BxPC3 cell proliferation. Our results suggest that circRNA_100782 may be a new marker and therapeutic target for PDAC.
Acknowledgment
This work was supported by Science and Technology Commission of Shanghai, China (No 13ZR1406700 and No 13DZ1930908).
Disclosure
The authors report no conflicts of interest in this work.
References
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. | ||
Hsu MT, Coca-Prados M. Electron-microscopic evidence for the circular form of rna in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. | ||
Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell. 2015;58(5):870–885. | ||
Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. | ||
Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. | ||
Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. | ||
Han D, Li J, Wang H, et al. Circular RNA MTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. | ||
Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. | ||
Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. | ||
Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. | ||
Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4 alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147(6):1233–1247. | ||
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. | ||
Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. | ||
Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. | ||
Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. | ||
Rebouissou S, Amessou M, Couchy G, et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457(7226):200–204. | ||
Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41(16):2502–2512. | ||
Scholz A, Heinze S, Detjen KM, et al. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology. 2003;125(3):891–905. | ||
Corcoran RB, Contino G, Deshpande V, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71(14):5020–5029. | ||
Huang C, Yang GA, Jiang T, Huang K, Cao J, Qiu Z. Effects of IL-6 and AG490 on regulation of Stat3 signaling pathway and invasion of human pancreatic cancer cells in vitro. J Exp Clin Cancer Res. 2010;29:51. | ||
Lesina M, Kurkowski MU, Ludes K, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–469. | ||
Xiao Y, Wang J, Yan W, et al. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting IL-6/STAT3 signalling. J Hepatol. 2015;62(4):889–896. | ||
Glazar P, Papavasileiou P, Rajewsky N. CircBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. | ||
Chen L, Zhang S, Wu J, et al. CircRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. | ||
Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8). | ||
Liu Y, Li PK, Li C, Lin J. Inhibition of STAT3 signaling blocks the anti-apoptotic activity of IL-6 in human liver cancer cells. J Biol Chem. 2010;285(35):27429–27439. | ||
Cheng Y, Li Y, Nian Y, Liu D, Dai F, Zhang J. STAT3 is involved in miR-124-mediated suppressive effects on esophageal cancer cells. BMC Cancer. 2015;15:306. | ||
Wang P, Chen L, Zhang J, et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33(4):514–524. | ||
Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. | ||
Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2016;388:208–219. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.