Back to Journals » Cancer Management and Research » Volume 13
CircRNA_0000429 Regulates Development of NSCLC by Acting as a Sponge of miR-1197 to Control MADD
Authors Wang JY, Zhang F, Hong L, Wei SJ
Received 22 August 2020
Accepted for publication 4 December 2020
Published 29 January 2021 Volume 2021:13 Pages 861—870
DOI https://doi.org/10.2147/CMAR.S270790
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Bilikere Dwarakanath
Jun-Yan Wang, Fan Zhang, Lei Hong, Su-Ju Wei
Department of Medical Oncology, Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei 050000, People’s Republic of China
Correspondence: Su-Ju Wei
Department of Medical Oncology, Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei 050000, People’s Republic of China
Tel +86 311-86095672
Email [email protected]
Background: Non-small-cell lung carcinoma (NSCLC) is the most common type of lung cancer. Circular RNA_0000429 (circ_0000429) is an identified circular RNA (circRNA) that is correlated with cancer progression. However, the role of circ_0000429 in NSCLC remains unknown. In the present study, we aimed to investigate the function of circ_0000429 in NSCLC and the underlying mechanism.
Methods: The expression patterns of circ_0000429 were determined using qRT-PCR in NSCLC samples and cell lines. The subcellular distribution of circ_0000429 in NSCLC cells was analyzed by fluorescence in situ hybridization (FISH). Cell proliferation was examined utilizing the CCK-8 assay. Cell migration and invasion were evaluated using the transwell assay. We used the bioinformatics software TargetScan and miRanda to predict circRNA–miRNA and miRNA–mRNA interactions.
Results: Our results showed that circ_0000429 expressions were significantly upregulated in NSCLC tissues and cell lines. Knockdown of circ_0000429 significantly inhibited the cell proliferation, migration, and invasion of NSCLC cells in vitro. Furthermore, we demonstrated that circ_0000429 acted as a sponge to absorb microRNA-1197 (miR-1197) and promoted MADD expression.
Conclusion: Collectively, our results demonstrated that circ_0000429 exhibited a carcinogenic role by sponging miR-1197 and regulating CMADD expression in NSCLC. These findings provided evidence for understanding the role of circ_0000429 in NSCLC tumorigenesis.
Keywords: circRNA_0000429, miRNA-1197, MADD, NSCLC
Introduction
In the cancer morbidity and mortality data released by GLOBOCAN2018, lung cancer is the most commonly diagnosed cause of cancer and cancer death in males and females.1,2 With the rapid growth of the population, the morbidity and mortality of lung cancer are getting higher and higher. Despite the development of new targeting and immunotherapy, the five-year survival rate of lung cancer is still very low.3 NSCLC accounts for about 80% of all lung cancer. The main pathological types are adenocarcinoma and squamous cell carcinoma.4 The current research focus is to explore the biological characteristics and pathogenesis of NSCLC and to find molecular markers for the diagnosis of NSCLC.
Circular RNA is a particular endogenous non-coding gene, which is covalently closed by reverse splicing of 3 ‘splice donor site and 5ʹ shear acceptor site to form a covalently closed loop.5,6 The expression level of highly conservative and stable in NSCLC, which is involved in the occurrence, development, and metastasis of NSCLC.7 It is expected to be an ideal biomarker. It can be used in the early diagnosis of NSCLC and provide new therapeutic targets and prognostic monitoring sites. Zhao et al collected 43 NSCLC patients' tissues and normal tissues and found that circRNA were up-regulated and 79 down-regulated. In NSCLC patients, the increased expression of circFADS2 was associated with advanced TNM stage, lymph node metastasis, poor differentiation, and poor overall survival.8 In the research of 149 cases of NSCLC and adjacent non-tumor tissues by J.WANG et al, they found that circ_0067934 was associated with the prognosis of NSCLC, and it was significantly correlated with TNM stage, lymph node status, distant metastasis, and overall survival. This gene acts as a tumor promoter in NSCLC and can inhibit epithelial-mesenchymal transformation when reducing the expression of circ_0067934. We know that epithelial-mesenchymal transformation (EMT) is significant for the invasion and metastasis of cancer cells. Circ_0067934 promotes the metastasis of NSCLC cells by regulating EMT. It is confirmed that the down-regulation of this gene inhibits the proliferation, migration, and invasion of NSCLC cells, indicating that it acts as an oncogene in the progression of NSCLC and may be a significant therapeutic target for NSCLC.9 Zhang H et al found that the expression of circZFR was markedly increased in NSCLC. Interfering of circZFR significantly prevented the cell progress of NSCLC cells. Furthermore, circZFR acted as a sponge of miR-101-3p and promoted cullin 4B expression.10 Tan S et al demonstrated that F-circEA-4a maybe a novel liquid biopsy biomarker for NSCLC.11 Circ_100,146 is highly expressed in NSCLC cell lines and tissue samples of NSCLC. Knockdown of circ_100146 inhibited the proliferation and invasion of NSCLC cells. Furthermore, circRNA 100146 could bind miR-361-3p and miR-615-5p to regulate multiple downstream mRNAs and further demonstrate endogenous competition among circ_100146, SF3B3, and miRNAs, providing novel insights into the mechanisms underlying NSCLC.12
It has been reported that circRNA could regulate the development and metastasis of NSCLC and act as an essential role in the prognosis of NSCLC, but the application of circRNA in targeted therapy of lung cancer is unclear. If we can further study, the use of circRNA on the regulation of NSCLC for targeted regulation and treatment should be a significant leap in the clinical treatment of NSCLC.
Here, we found that circRNA_0000429 was markedly increased in 56 NSCLC samples compared with their paired adjacent normal tissues by RT-PCR. Interfering expression of circRNA_0000429 inhibited the proliferation and metastasis in NSCLC cell lines by controlling the expression of miR-1197. The silencing of miR-1197 would disturb the inhibition function of circRNA_0000429 on NSCLC proliferation and development. These findings contribute to an understanding of the processes involved in migration. Further, circRNA_0000429 could induce the expression of MADD by sponging miR-1197. In conclusion, we revealed that circRNA_0000429 could act as a ceRNA to regulate the expression of MADD by binding with miR‐1197.
Materials and Methods
Clinical Samples
The tumor samples and adjacent normal samples were collected from 56 NSCLC patients at Fourth Hospital of Hebei Medical University (Table 1). All of the patients provided their written informed consent to participate in this study. This study was reviewed and approved by the Ethics Committee of Fourth Hospital of Hebei Medical University, which was conducted according to the international guidelines of the Helsinki Declaration.
 |
Table 1 Clinicopathological Features of NSCLC Patients (N= 56) |
Cell Culture
Beas-2B, H2170, A549, SPC-A-1, PG49 cell lines were purchased from the Science Cell Laboratory. Cell lines were cultured in PRIM 1640 with 10% FBS and 100 μL/mL penicillin and streptomycin (Beyotime, China) and placed at 37°C with 5% CO2.
Quantitative Real-Time PCR
The tissue or cells were taken, and total RNA was extracted according to the operating instructions of Rneasy Mini Kit. Total RNA concentration was determined by spectrophotometer. Then, according to the operation instructions of the reverse transcription kit, the extracted RNA was reverse-transcribed into cDNA and stored at −80°C for later experiments. Quantitative PCR reaction system was configured according to the instructions. The reaction conditions were as follows: 9°C 15min, 40 cycles, 95°C 15s in each cycle, 55°C 30s, 72°C 1s, 40°C 1min. The data were calculated by 2−ΔΔCt. The data were calculated by 2-ΔΔCt. The primer sequence was shown as follows. Circ_0000429 Forward: CCTTAGTCCCAATGCCTCCG. Reverse: ACCCGCACACGGAAAACTTA. GAPDH Forward: GGTGA AGGTC GGAGT CAACG. Reverse: CAAAG TTGTCATGGATGHACC.
Western Blot
Total protein was collected from tissues or cells with RIPA lysis Mix. The Western blotting assay was performed as previously described. Briefly, 50–80 μg protein extraction was loaded via SDS-PAGE and transferred onto nitrocellulose membranes (Absin, China); then, incubated with primary antibodies for 2 hrs at temperature, then plated at 4°C overnight, the membranes were incubated in 5% non-fat milk blocking buffer for horizontal mode 3 h. After incubation with secondary antibodies, the membranes were scanned using an Odyssey, and data were analyzed with Odyssey software (LI-COR, USA).
CCK8 Assay
Cells were cultured in 96-well cell plates and added CCK-8 buffer (MedChemExpress, USA) at 0, 24, 48, and 72 h. 2 h later, measure 450 OD value with an MK3ELISA photometer (Tecan, Germany).
In vivo Tumor Growth Assay
Nude mice were purchased from the Beijing Charles river. After creating low expression and normal expression of circ_0000429 in A549, A549 cells were subcutaneously injected in the right lower limb of the nude mice. Tumor size was measured every five days. After another 15 d of injection, mice were intraperitoneally injected with 3% pentobarbital sodium and were euthanatized by excessive anesthesia with a dose of 90 mL/kg, and the tumors were removed for follow-up study. The research protocol of this study was approved by the Animal Care and Use Committee of Fourth Hospital of Hebei Medical University and was performed based on the guidelines and regulation of the Management of Laboratory Animals published by the Ministry of Science and Technology of the People’s Republic of China. All animal experiments took place at Fourth Hospital of Hebei Medical University.
Cell Apoptosis Assay
The cells were counted, about 5×105 cells/mL. Then, 1 mL cells were centrifuged, 1000 rpm, 10 min, 4°C, and the medium was thrown away. The cells were washed with PBS and dropped medium. The cells were resuspended and avoid light for 15 min, 200 μL Binding Buffer with10 μL Annexin V-FITC, and 10 μL PI. Flow cytometry was used to measure apoptosis rate within 1 h.
Cell Cycle Assay
A549 cells were collected with 1mL trypsin for 2min, suspension the cell with 5mL PBS, centrifuge at 1000 RPM for 5 min at 4°C. 10mL PBS buffer was used to the re-washed and dropping medium; Then, the cells were fixed with 70% ethanol overnight. The next day, the cell medium was filtered with a 300-mesh sieve, centrifuged at 1000 RPM at 4°C for 5min, and the supernatant was discarded. The cells were avoided light and fixed with 1mL PI solution and stated at 4°C for 30 min. Flow cytometer was used to evaluate the cell cycle.
Luciferase Assay
20 mmol/L miR-1197 mimic or miR-NC, and circ_0000429-WT/circ_0000429-mutation or MADD-WT/MADD-mutation were co-transfected into HEK293T cells. Luciferase activity was detected with Luciferase Reporter Assay Kit (Biovision, China) on luminometer (Berthold, Germany) 48 hr after the transfection.
Statistical Analysis
All values are calculated as the mean ± SEM. Statistical significances were measured by Student’s t-test and ANOVA. A two-tailed value of P < 0.05 was indicated as a statistically significant difference. Data statistics were used the GraphPad 7.0.
Results
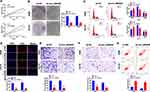
CircRNA_0000429 is Up-Regulated in NSCLC
To detect the function of circRNA_0000429 in NSCLC, we collected the 56 NSCLC patients’ tumor tissues and normal lung tissues. After detecting by RT-PCR, we observed that circRNA_0000429 was up-regulated in tumor tissues (Figure 1A). Meanwhile, we determined the expression of circRNA_0000429 in five NSCLC cell lines (A549, H2170, SPC-A-1, PG49). Beas-2 B was indicated as a control. The results showed an increased level of circRNA_0000429 in NSCLC cell lines (Figure 1B). The abnormal expression of circRNA_0000429 may play an important role in NSCLC, which needs us further exploration.
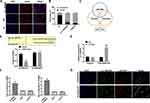
Knockdown of circRNA_0000429 Prevents Proliferation and Metastasis in NSCLC Cell Lines
Next, we constructed the shRNA for decreasing the level of circRNA_0000429, and sh-NC was indicated as a negative control. CCK8 was performed to assess the cell viability of A549 and H2170 cells. Forced decreased expression of circRNA_0000429 markedly inhibited the cell viability both in NSCLC cells (Figure 2A). Colony formation assay revealed that sh-circRNA_0000429 prevented cell proliferation in NSCLC cells (Figure 2B). By analyzing the cell cycle, we found that sh-circRNA_0000429 prevented the cells from G1/G0 phase into the S phase (Figure 2C). As Figure 2D shown, knockdown of circRNA_0000429 remitted the proliferation of NSCLC cells by employing the EdU (5-Ethynyl-2ʹ-deoxyuridine) assay. Further, sh-circRNA_0000429 significantly prevented the migration and invasion in A549 and H2170 cells (Figure 2E and F). The results of flow cytometry confirmed that sh-circRNA_0000429 induced the apoptosis in NSCLS cells (Figure 2G). Taken together, knockdown of circRNA_0000429 would prevent proliferation and metastasis, induced apoptosis in A549 and H2170 cells.
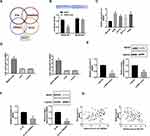
CircRNA_0000429 Indicates as a Sponge for miRNA-1197
To deeply detect the molecular mechanisms of circRNA_0000429 in NSCLC, we detected the subcellular location of circRNA_0000429. CircRNA_0000429 is predominantly located in the cytoplasm of A549 cells by performing FISH and separation of cytoplasm and nucleus fraction RNA analyses (Figure 3A and B). As previous reports showed, circRNA_0000429 could as a sponge of miRNA-1197 and regulate the level of miRNA-1197. Bioinformatics website predicted the existence of binding sites between circRNA_0000429 and miRNA-1197. Next, we performed luciferase assays to confirm whether circRNA_0000429 binds to miRNA-1197. The assay report showed that circRNA_0000429 could bind with miRNA-1197 (Figure 3C and D). Further, we carried out anti-AGO2 RIP in A549 cells. The Endogenous circRNA_0000429 pull-down by AGO2 was enriched in cells transfected with miRNA-1197 (Figure 3E). To investigate the function of miRNA-1197 with circRNA_0000429, a circRNA-specific probe was used to pull down its interactive miRNA-1197s. circRNA_0000429 precipitation complex performed the enrichment of miRNA-1197 (Figure 3F). Therefore, the relationship of circRNA and miRNA-1197 was verified by FISH assay, and we found that miRNA-1197 was co-localized with circRNA_0000429 in the cytoplasm, which once performed a relationship between circRNA_0000429 and miRNA-1197 (Figure 3G). In summary, circRNA_0000429 could interact with miRNA-1197 as a sponge.
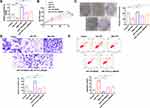
MiRNA-1197 Could Bind with the 3ʹUTR of MADD
Bioinformatics website predicted the existence of binding sites between 3ʹUTR of MADD and miRNA-1197. The luciferase assay report showed that miRNA-1197 could bind with MADD (Figure 4A and B). Meanwhile, we determined the level of MADD was also markedly increased in different NSCLC cell lines (Figure 4C). Biotin pull‐down assays indicated that the miRNA-1197 was distinctly enriched with MADD (Figure 4D). RT-PCR assay results revealed that forced expression of miRNA-1197 decreased the level of MADD (Figure 4E). Knockdown of circRNA_0000429 also prevented the expression of MADD (Figure 4F). A positive relationship between circ_0000429 and MADD was found in the NSCLC patient tissue samples. In addition, using RT-qPCR, we measured the expression of miR-1197 in the 56 NSCLC patient tumor tissues. A negative relationship between MADD and miR-1197 was found in the NSCLC patient tissues (Figure 4G).
CircRNA_0000429 Involves in Tumor Progress by Regulating miRNA-1197/MADD
Based on the above relationship research of circRNA_0000429, miRNA-1197 and MADD, we co-cultured the miRNA-1197, AMO-miRNA-1197, MADD and circRNA_0000429 into A549 cells, the expression of MADD was detected in every group. MiRNA-119 prevented the expression of MADD, and co-cultured with MADD or circRNA_0000429 recovered the level of MADD, which was similar to AMO-miRNA-1197 (Figure 5A). Next, we assessed the cell viability of A549 cells after co-cultured. Overexpression of c6ircRNA_0000429 or MADD would abolish the proliferation function of miRNA-1197 (Figure 5B). Colony formation assay performed similar results (Figure 5C). By employing the transwell assay, overexpression of miRNA-1197 inhibited the invasion of A549 cells, which was blocked by co-culturing with circRNA_0000429 or MADD (Figure 5D). MiRNA-1197-inducing apoptosis would be prevented by circRNA_0000429 or MADD (Figure 5E). Taken together, circRNA_0000429/miRNA-1197/MADD axis participated in the growth and metastasis of A549 cells.
Knockdown of CircRNA_0000429 Prevents Tumorigenesis in vivo
Base on the above results, we constructed stable low expression of circRNA_0000429 A549 cell (sh-circRNA_0000429-A549), normal level of circRNA_0000429 A549 cell as a control (sh-NC-A549), Two groups of A549 cells were randomly and subcutaneously injected in the right lower limb of the nude mice. Tumor size was measured every five days. Sh-circRNA_0000429-A549 injection group significantly reduced tumor volume and weight (Figure 6A–C). The tumor tissues of sh-circRNA_0000429-A549 injection group showed a higher level of miRNA-1197, lower level of circRNA_0000429 and MADD compared with sh-NC-A549 (Figure 6D and E). H&E staining showed the morphological changes for the tumor cell growth between sh-NC and sh-circRNA_0000429 groups, and immunohistochemistry (IHC) analysis indicated the decreased expression levels of Ki-67 and MADD (Figure 6F). To verify the in vitro findings, we examined the biological role of circ_0000429 in mediating in vivo metastasis. A549 cancer cells with stably decreased expression of circ_0000429 expression were subcutaneously implanted into nude mice. Consistent with the above in vitro findings, the downregulation of circ_0000429 dramatically inhibited lung metastasis (Figure 6G).
Discussion
CircRNA plays a related role in the occurrence, metastasis and prognosis of lung cancer, the abnormal expression of circular RNA may be triggered by healthy cells to cancer cells, or it may be the result of protein or enzyme imbalance in cancer cells.13,14 However, further study is still needed to track problems that can be treated on cell regulation, including gene mutations, DNA methylation, cell cycle disorders, and changes in protein or enzyme activity. The latest experimental study shows that the expression level of annular RNA in NSCLC tissues is significantly higher than that in normal tissues, and its expression profile can become a potential target for early diagnosis of NSCLC and improve the accuracy of early diagnosis.15,16 In terms of treatment, circular RNA can provide more accurate therapeutic targets, achieve more accurate therapeutic effects, and reduce damage to normal tissues and cells. At the same time, the abnormal expression of circular RNA affects the growth, development and metastasis of NSCLC cells, which is closely related to lymph node metastasis and tumor stage, which can accurately judge the stage and prognosis of lung cancer.
Zhao et al found that there are 356 circular RNA expression differences between non-small cell adenocarcinoma and adjacent normal tissues, which reveals the circular RNA expression profile of early lung adenocarcinoma, which provides a potential target for the diagnosis of early lung adenocarcinoma, and provides a new idea for the new gene diagnosis of NSCLC.17 Yao et al revealed that circRNA100876 was significantly up-regulated in NSCLC tissues compared with adjacent non-tumor cells. Statistics show that the expression of circRNA100876 has far-reaching significance for tumor invasion and metastasis.18 Luo et al found the increased level of hsa_circ_0000064 in lung cancer, and its abnormal expression was related to T stage, lymphatic metastasis and TNM stage, which would be a kind of new potential biomarkers of lung cancer.19 Zhu et al first found that hsa_circ_0013958 was up-regulated in all lung adenocarcinoma tissues, cells and plasma, and its level was related to TNM stage and lymphatic metastasis.20 Zhang PF et al reported that circFGFR1 promoted the migration, invasion, proliferation, and immune evasion of NSCLC cells by sponging miR-381-3p and regulating target gene C-X-C motif chemokine receptor 4 (CXCR4), which promoted NSCLC progression and resistance to anti-programmed cell death 1 (PD-1)-based therapy.21 Therefore, circRNA plays a vital function in early diagnosis, therapy and prognosis of NSCLC. In our study, we firstly revealed the role of circ_0000429 in NSCLC. The forced decreased expression of circ_0000429 would inhibit the ability of proliferation, migration, invasion and induced apoptosis. The results showed that miR-1197 expression was obviously downregulated in both NSCLC tissues and cell lines. Furthermore, we found that circ_0000429 executes its carcinogenic role in NSCLC by sponging miR-1197.
In previous research, MADD is a cancer cell pro-survival factor, which would regulate the anti-tumor activity of TRAIL in anaplastic thyroid cancer.22 Silencing of MADD prevented migration, invasion, clonogenic capacity, mitochondrial length and potential via controlling EMT and Wnt signaling pathway.23 Gao P et al revealed that circ_0074027/BRD4/MADD involved in NSCLC progression.24 Meanwhile, MADD is a downstream target of PTEN in triggering apoptosis.25 Further investigations revealed that miR-1197 overexpression-caused decreases in cell proliferation, migration and invasion were significantly prevented by overexpression of MADD. The results indicated that miR-1197 exerted its tumor suppresser effects by targeting MADD in NSCLC.
At present, the research results on circular RNA and its functions are still very limited. With the continuous emergence of new methods and technologies, this problem will be fully solved by medical and scientific researchers.
Conclusions
Collectively, our study showed that circ_0000429 was upregulated in NSCLC tissues and cell lines. Knockdown of circ_0000429 in NSCLC cells significantly inhibited cell proliferation, migration and invasion. Furthermore, we demonstrated that circ_0000429 executed a carcinogenic role by regulating miR-1197/MADD axis.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors declare no competing interests.
References
1. Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. J Nat Rev Cancer. 2019;19(9):495–509. doi:10.1038/s41568-019-0179-8
2. Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. J JAMA. 2019;322(8):764–774. doi:10.1001/jama.2019.11058
3. Cheng H, Perez-Soler R. Leptomeningeal metastases in non-small-cell lung cancer. J Lancet Oncol. 2018;19(1):e43–e55. doi:10.1016/S1470-2045(17)30689-7
4. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
5. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881.e813. doi:10.1016/j.cell.2018.12.021
6. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
7. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. J Nat Rev Genet. 2016;17(11):679–692. doi:10.1038/nrg.2016.114
8. Zhao F, Han Y, Liu Z, et al. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38(4):38. doi:10.1042/BSR20180570
9. Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22:3053–3060.
10. Zhang H, Wang X, Hu B, et al. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells, Nanomed Biotechnol. 2019;47(1):3410–3416. doi:10.1080/21691401.2019.1652623
11. Tan S, Sun D, Pu W, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. 2018;17(1):138. doi:10.1186/s12943-018-0887-9
12. Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):13. doi:10.1186/s12943-019-0943-0
13. Van Der Steen N, Lyu Y, Hitzler AK, et al. The circular RNA landscape of non-small cell lung cancer cells. Cancers. 2020;12(5):12. doi:10.3390/cancers12051091
14. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. J Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
15. Nicot C. RNA-Seq reveal the circular RNAs landscape of lung cancer. J Mol Cancer. 2019;18(1):183. doi:10.1186/s12943-019-1118-8
16. Hall IF, Climent M, Quintavalle M, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. 2019;124(4):498–510. doi:10.1161/CIRCRESAHA.118.314240
17. Zhao J, Li L, Wang Q, et al. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44(6):2138–2146. doi:10.1159/000485953
18. Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi:10.1016/j.prp.2017.02.011
19. Luo YH, Zhu XZ, Huang KW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi:10.1016/j.biopha.2017.12.015
20. Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi:10.1111/febs.14132
21. Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer. 2019;18(1):179. doi:10.1186/s12943-019-1111-2
22. Saini S, Sripada L, Tulla K, et al. MADD silencing enhances anti-tumor activity of TRAIL in anaplastic thyroid cancer. Endocr Relat Cancer. 2019;26(6):551–563. doi:10.1530/ERC-18-0517
23. Fasolo F, Di Gregoli K, Maegdefessel L, Johnson JL. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc Res. 2019;115(12):1732–1756. doi:10.1093/cvr/cvz203
24. Navasiolava N, Degryse B, Custaud MA, Moyna NM, Murphy RP. Deciphering the mechanisms behind cardiovascular disease: long noncoding RNAs as key molecular signaling hubs and biomarkers of atherosclerosis. J Cardiovasc Pharmacol. 2020;76(2):125–127. doi:10.1097/FJC.0000000000000863
25. Jayarama S, Li LC, Ganesh L, et al. MADD is a downstream target of PTEN in triggering apoptosis. J Cell Biochem. 2014;115:261–270.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.