Back to Journals » OncoTargets and Therapy » Volume 13
CircRNA ZNF609 Knockdown Suppresses Cell Growth via Modulating miR-188/ELF2 Axis in Nasopharyngeal Carcinoma
Received 10 October 2019
Accepted for publication 16 January 2020
Published 23 March 2020 Volume 2020:13 Pages 2399—2409
DOI https://doi.org/10.2147/OTT.S234230
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Mingyan Li, Yujie Li, Min Yu
Department of Otolaryngology-Head and Neck Surgery, Zhengzhou Central Hospital Affiliated to Zhengzhou University, Zhengzhou, People’s Republic of China
Correspondence: Mingyan Li
Department of Otolaryngology-Head and Neck Surgery, Zhengzhou Central Hospital Affiliated to Zhengzhou University, No. 16, Tongbai North Road, Zhongyuan District, Zhengzhou, Henan 450007, People’s Republic of China
Tel +86-0371-67690918
Email [email protected]
Background: Circular RNAs (circRNAs) and microRNAs (miRNAs) have been reported to act as the important regulators in nasopharyngeal carcinoma (NPC). CircRNA ZNF609 (circ-ANF609) and miR-188 have been, respectively, reported to play a pro-cancer and anti-cancer role in NPC. The purpose of this study is to reveal the functional relation of circ-ZNF609 and miR-188 in NPC development.
Methods: The transcription level and protein level of genes were detected by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot assay, respectively. Cell proliferation was analyzed using 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Furthermore, flow cytometry analysis was used to assess cell cycle transition and cell apoptosis rate. Besides, the interaction between miR-188 and circ-ZNF609 or E74-like factor 2 (ELF2) was predicted by starbase or microT-CDS, and then confirmed by the dual luciferase reporter assay and RIP assay.
Results: Circ-ZNF609 and ELF2 levels were increased and miR-188 level was decreased in NPC. Circ-ZNF609 knockdown significantly inhibited cell proliferation and cell cycle transition, as well as accelerated apoptosis in NPC cells. Interestingly, circ-ZNF609 directly bound to miR-188. Circ-ZNF609 regulated NPC cell growth through modulating miR-188 expression. In addition, miR-188 suppressed NPC cell growth via directly targeting ELF2. Finally, we confirmed that circ-ZNF609 mediated miR-188 level to modulate ELF2 expression.
Conclusion: Our findings demonstrated that circ-ZNF609 depletion-repressed proliferation and cell cycle transition, and induced apoptosis of NPC cells via modulation of miR-188/ELF2 axis, providing potential targets for the therapy of NPC.
Keywords: CircRNA ZNF609, MiR-188, ELF2, cell growth, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC), one of the head and neck cancers, is a malignancy that is the most prevalent epithelial cancer in adults and mainly occurs in Asian and Northern Africa.1 According to statistics in 2018, the 5 years survival rate of NPC was less than 70%.2 Nowadays, Radiation therapy is the main strategy for the therapy of NPC patients, whereas radio-resistance decreases the treatment effect.3 Therefore, it is essential to explore the mechanism of NPC development for the therapy of NPC patients.
In recent years, non-coding RNAs, including long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs), were discovered.4,5 LncRNAs and miRNAs were reported to exert function and considered as the biomarkers in NPC.6–9 Compared with them, the functional mechanism of circRNAs was less studied. Present studies suggested that circRNAs, with a circular configuration, were involved in the translation regulation of genes and the development of human cancers.10–12 CircRNA ZNF609 (circ-ZNF609) was identified as a circRNA that located at chr15:64791491-64792365. Accumulating evidence indicated that circ-ZNF609 was a positive regulator for cancer development. For example, Wu et al demonstrated that circ-ZNF609 enhanced colorectal cancer cell motility via regulating miR-150/Gli1 axis.13 Wang et al indicated that circ-ZNF609 promoted cell proliferation and invasion through regulation of miR-145-5p and p70S6K1 in breast cells.14 Furthermore, circ-ZNF609 expression was increased and circ-ZNF609 accelerated cell growth through modulating miR-150-5p in NPC cells.15 Therefore, circ-ZNF609 plays a pivotal role in human cancers containing NPC. The study of ZNF609 function is needed for the treatment of NPC.
MicroRNAs (miRNAs), identified as the small non-coding RNAs, consist of approximately 20 nucleotides and play crucial roles in human diseases through modulating gene translation and mRNA degradation.16 In the past few decades, amounting reporters confirmed that miRNAs exerted function in various types of cancer cell progression, including proliferation, invasion, apoptosis, and autophagy.17–19 Besides, it is reported that miRNAs are related to drug resistance.20 According to the prediction, estimated 60% of genes are regulated by miRNAs in mammals.21 MiR-188, an endogenous miRNA, was first reported in 2013.22 This paper indicated that miR-188 regulated synaptic transmission and plasticity as well as its expression was increased under the induction of long-term potentiation condition. Thereafter, miR-188 was reported to modulate cell senescence in bone marrow and suppress the proliferation and cell cycle in glioma.23,24 Also, miR-188 played an important function in NPC. For example, Wu et al suggested that miR-188 inhibited G1/S transition through regulating cyclin/CDK axis in NPC cells.25 However, the study of miR-188 in NPC is rare. Therefore, it is necessary to explore the functional mechanism of miR-188 in NPC.
E74-like factor 2 (ELF2), identified as a transcription factor, is reported to regulate gene expression through associating with RUNX1.26 Previous evidence demonstrated that the genes interacted with ELF2 was related to lymphocyte function.27 Besides, ELF1 and ELF4, two members of ELF subfamily, are reported to mediate T cell growth-related genes and exert function in natural killer cells.28–30 Nowadays, increasing studies of ELF2 function were carried out, and confirmed that ELF2 was involved in cancer development. Zhang et al revealed that ELF2 promoted the proliferation of osteosarcoma cells.31 Jin et al suggested that ELF2 was considered as a potential target for the prognosis of non-small cell lung cancer.32 Besides, ELF2 was highly expressed in NPC tissues and ELF2 upregulation promoted the proliferation of NPC cells.33 However, the functional mechanism of ELF2 is rarely studied in NPC.
Here, we detected the levels of circ-ZNF609, miR-188, and ELF2, and analyzed circ-ZNF609 function through downregulating circ-ZNF609 expression in NPC. Furthermore, the function of miR-188 and ELF2 in circ-ZNF609-regulated NPC cell progression was explored.
Materials and Methods
Tissue Samples and Cell Culture
50 NPC tissues and 50 corresponding normal paracancerous tissues were obtained from NPC patients subjected to surgical resection at Zhengzhou Central Hospital Affiliated to Zhengzhou University, and then frozen in liquid nitrogen for further experiment. This study was approved by the Ethics committee of Zhengzhou Central Hospital Affiliated to Zhengzhou University. All patients signed informed consent and did not receive any therapy before this assay.
Normal cells (NP69, JN-3146) and NPC cells (C666-1, JN-E1537 and 5-8F, JN-3176) were provided by Jining Shiye (Shanghai, China). C666-1 and 5-8F cells were maintained in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA, A1049101) containing 10% fetal bovine serum (FBS; Gibco, 10099141) while NP69 cells were cultured in serum-free medium Keratinocyte-SFM (Gibco, 10744019) at 37°C under 5% CO2 condition.
Plasmid and Transfection
Si-circ-ZNF609 and negative control (si-con) were obtained from Ribobio (Guangzhou, China). For overexpression of circ-ZNF609 and ELF2, their sequences were, respectively, amplified and cloned into the pcDNA3.1 (+) CircRNA Mini and pcDNA3.1 plasmid (V79020) from Invitrogen (Carlsbad, CA, USA). MiR-188 mimics and miRNA inhibitors (in-miR-188) were synthesized and purified by GenePharma (Shanghai, China).
Cell transfection assay in this study was carried out using Lipofectamine 2000 (Invitrogen, 11668019) based on the user’s manual.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was obtained using TRIzol (Invitrogen, 15596018) based on the user’s manual and then used to synthesize cDNA using SuperScript II first-strand synthesis system (Invitrogen, 11904018). QRT-PCR was carried out using SYBR Green qPCR Master Mix (Takara, Dalian, China, RR820L/W). Gene expression was normalized against U6 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH). 2−ΔΔCT method was applied for the analysis of gene expression. Primers for miR-188 (#A25576) were acquired from Applied Biosystems (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The primer sequences against circ-ZNF609, ELF2, U6 and GAPDH were: circ-ZNF609 forward (F), 5ʹ-CAGCGCTCAATCCTTTGGGA-3ʹ and reverse (R), 5ʹ-GACCTGCCACATTGGTCAGTA-3ʹ; ELF2 F, 5ʹ-GAGTGCGTGAGTGTGTGTCG-3ʹ and R, 5ʹ-TGAACTGATGCTTCCACGGC-3ʹ; U6 F, 5ʹ-TGCGGGTGCTCGCTTCGGCAGC-3ʹ and R, 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ; GAPDH F, 5ʹ-CATGAGAAGTATGACAACAGCCT-3ʹ and R, 5ʹ-AGTCCTTCCACGATACCAAAGT-3ʹ.
Cell Proliferation Assay
In this research, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was conducted to assess cell proliferation in line with the recommended protocol (Invitrogen, M6494). Briefly, 5-8F and C666-1 cells were transfected with vectors and incubated for 0 h, 24 h, 48 h or 72 h, respectively. Subsequently, the cells were treated with 20 µL MTT solution for 4 h first, and then the supernatant was removed. Next, 100 µL dimethyl sulfoxide was added to dissolve products. Finally, a microplate reader (Bio-Rad, Richmond, CA, USA, 168-1000XC) was used to measure the absorbance under 490 nm condition.
Cell Apoptosis Assay
The apoptosis rate of 5-8F and C666-1 cells was analyzed with propidium iodide (PI)/Annexin V Cell Apoptosis Kit (Invitrogen, V13245) in this assay based on the user’s manual. In brief, 5-8F and C666-1 cells were transfected with vectors and then cultured for 48 h. Next, FITC-Annexin V and PI were applied to stain cells. Finally, cell apoptosis rate was examined by flow cytometry (FACSCantoII, 338960; BD Biosciences, San Jose, CA, USA).
The Dual Luciferase Reporter Assay
Firstly, circ-ZNF609-WT/MUT and ELF2-WT/MUT were constructed via cloning their sequence into pGL3 vector (Promega, Madison, WI, USA, E1751). Then, every construction was co-transfected into 5-8F and C666-1 cells with miR-188 or miR-con. Finally, luciferase density was examined by the Dual Luciferase Reporter Assay System (Promega, E1910).
RNA Immunoprecipitation (RIP) Assay
RIP assay in this research was carried out with Magna RNA immunoprecipitation kit (Millipore, Billerica, MA, USA, 17-700) according to the user’s protocol. Briefly, 5-8F and C666-1 cells were cultured for 48 h, and lysed by RIP buffer containing magnetic beads conjugated with Ago2 or IgG antibody. Then, TRIzol reagent was used to collect immunoprecipitated RNAs and enrichment of circ-ZNF609 or ELF2 was analyzed using qRT-PCR assay.
Western Blot Assay
Briefly, NPC cells were collected and then lysed using lysis buffer for extraction of total proteins. Next, the proteins were separated on dodecyl sulfate, sodium salt-Polyacrylamide gel electrophoresis (SDS-PAGE, 10%), transferred to PVDF members (Millipore, IPVH00010), and incubated using primary antibodies (1:1000; Abcam, Cambridge, MA, USA) against ELF2 (ab225958) and β-actin (ab8227). Finally, the proteins were incubated with the second antibodies (Abcam, ab205718) and analyzed by the ECL plus Kit (Beyotime, Haimen, China, P0018S).
Statistical Analysis
Student’s t-test or one-way ANOVA was performed for the analysis of significant difference (P< 0.05). Data were represented as the means ± standard deviation (SD) and obtained under at least three independent biological reduplications condition.
Results
Circ-ZNF609 Expression Was Increased in NPC Tissues and Cells
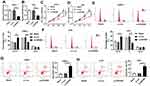
Firstly, we detected the expression level of circ-ZNF609 in NPC tissues and normal tissues, and found that circ-ZNF609 expression was higher in NPC tissues than that in normal tissues (Figure 1A). Then, circ-ZNF609 level in NPC cells (C666-1 and 5-8F) and normal cells (NP69) was also analyzed. As shown in Figure 1B, circ-ZNF609 level was significantly upregulated in NPC cells. Therefore, circ-ZNF609 might play an important role in NPC development.
Circ-ZNF609 Knockdown Suppressed Cell Proliferation and Cycle, and Induced Apoptosis in NPC Cells
To investigate the function of circ-ZNF609 in NPC cells, the effect of circ-ZNF609 knockdown on NPC cell growth was explored. Firstly, C666-1 and 5-8F cells were transfected with si-con or si-circ-ZNF609. QRT-PCR analysis suggested that the transfection of si-circ-ZNF609 remarkably downregulated circ-ZNF609 level (Figure 2A and B). Then, MTT assay was performed to assess cell proliferation. The results suggested that circ-ZNF609 depletion dramatically suppressed cell proliferation (Figure 2C and D). Next, we analyzed the percentage of cells in different cycle stages, including G0-G1, S, and G2-M. As indicated in Figure 2E and F, circ-ZNF609 knockdown inhibited cell cycle transition from G0-G1 stage to S stage in C666-1 and 5-8F cells. Besides, increased cell apoptosis was observed in circ-ZNF609-depleted NPC cells (Figure 2G and H). These results indicated that the downregulation of circ-ZNF609 repressed cell proliferation and cycle, and accelerated apoptosis in NPC cells.
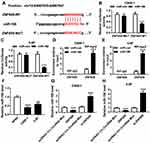
Circ-ZNF609 Served as a Sponge for miR-188
Next, we predicted the potential target genes of circ-ZNF609 using bioinformatics analysis tool starbase, and found that miR-188 likely interacted with circ-ZNF609 (Figure 3A). Then, the dual luciferase reporter assay was performed to confirm this possibility. As demonstrated in Figure 3B and C, miR-188 significantly reduced luciferase activity of cells transfected with circ-ZNF609-WT, but not circ-ZNF609-MUT, revealing that circ-ZNF609 directly bound to miR-188. Meanwhile, this interaction was also confirmed by RIP assay in NPC cells (Figure 3D and E). Moreover, we found that miR-188 expression was decreased in NPC cells (Figure 3F). Finally, the effect of circ-ZNF609 on miR-188 expression was investigated. The results demonstrated that circ-ZNF609 overexpression downregulated miR-188 expression, whereas circ-ZNF609 knockdown strongly increased miR-188 level in C666-1 and 5-8F cells (Figure 3G and H). Thus, circ-ZNF609 directly downregulated miR-188 expression in NPC cells.
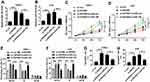
MiR-188 Knockdown Reversed the Effect of circ-ZNF609 Depletion on NPC Cell Growth
Based on the above results, it was speculated that circ-ZNF609 knockdown inhibited NPC cell growth via regulating miR-188 expression. To confirm this hypothesis, C666-1 and 5-8F cells were transfected with si-con, si-circ-ZNF609, si-circ-ZNF609+ in-miR-con, or si-circ-ZNF609+ in-miR-188, respectively. QRT-PCR analysis suggested that miR-188 expression was upregulated by the transfection of si-circ-ZNF609, and then downregulated by the transfection of in-miR-188 (Figure 4A and B). Next, cell proliferation in these cells was assessed. As shown in Figure 4C and D, cell proliferation was suppressed by circ-ZNF609 knockdown, whereas this change was weakened by decreased miR-188 expression. In addition, the inhibitory effect of miR-188 depletion on circ-ZNF609 knockdown-regulated cell cycle transition was observed in NPC cells (Figure 4E and F). Furthermore, we analyzed cell apoptosis rate, and found that miR-188 knockdown reversed the effect of circ-ZNF609 depletion on NPC cell apoptosis (Figure 4G and H). These data demonstrated that circ-ZNF609 regulated cell development through modulating miR-188 expression.
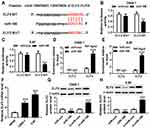
ELF2 Was a Target Gene of miR-188
Bioinformatics analysis website microT-CDS predicted that ELF2 was a potential target of miR-188 (Figure 5A). To verify this prediction, the dual luciferase reporter assay was carried out. The data demonstrated that the cells co-transfected with miR-188 and ELF2-WT exhibited lower luciferase activity than the cells co-transfected with miR-188 and ELF2-MUT (Figure 5B and C), confirming the interaction between miR-188 and ELF2. Also, this interaction was also verified by RIP assay in NPC cells (Figure 5D and E). In addition, increased ELF2 level was observed in NPC cells (Figure 5F). Besides, whether miR-188 modulated ELF2 expression was analyzed in NPC cells. As shown in Figure 5G and H, miR-188 upregulation significantly decreased ELF2 level, and miR-188 knockdown dramatically increased ELF2 level in C666-1 and 5-8F cells. These data indicated that miR-188 negatively regulated ELF2 expression through interaction in NPC cells.
MiR-188 Repressed Cell Growth Through Regulating ELF2 Expression in NPC Cells
To investigate whether miR-188 regulated ELF2 expression to repress NPC cell growth, C666-1 and 5-8F cells were transfected with miR-con, miR-188, miR-188 + pcDNA3.1(+), or miR-188 + ELF2, respectively. Then, qRT-PCR assay was performed to detect the expression level of ELF2. As indicated in Figure 6A and B, miR-188 overexpression significantly decreased ELF2 level, and then ELF2 level was partly rescued by overexpressing ELF2. Next, the effect of ELF2 on miR-188-mediated cell proliferation was analyzed. From Figure 6C and D, the upregulation of ELF2 induced cell proliferation suppressed by miR-188 overexpression. Also, we confirmed that miR-188 overexpression repressed cell transition, whereas this action was reversed by increased ELF2 level in NPC cells (Figure 6E and F). Besides, that ELF2 upregulation weakened the effect of miR-188 overexpression on cell apoptosis rate was observed in C666-1 and 5-8F cells (Figure 6G and H). Taken together, miR-188 inhibited cell growth by modulation of ELF2 level in NPC cells.
Circ-ZNF609 Mediated miR-188 Expression to Modulate ELF2 Level
Finally, we supposed that circ-ZNF609 regulated miR-188 expression to affect ELF2 level. To verify this speculation, C666-1 cells were transfected with pcDNA3.1(+) CircRNA Mini, circ-ZNF609, circ-ZNF609 + miR-con, or circ-ZNF609 + miR-188, respectively, and the levels of miR-188 and ELF2 were detected. As demonstrated in Figure 7A and C, the transfection of circ-ZNF609 decreased miR-188 level and increased ELF2 level, as well as the transfection of miR-188 partly rescued miR-188 level and reduced ELF2 level. Meanwhile, this speculation was also confirmed by transfecting si-con, si-circ-ZNF609, si-circ-ZNF609+ in-miR-con, or si-circ-ZNF609 + in-miR-188 into 5-8F cells, respectively (Figure 7B and D). Therefore, circ-ZNF609 downregulated miR-188 expression to increase ELF2 level.
Discussion
In recent years, circRNA/miRNA/mRNA signaling pathways have been reported to involve the development of human cancers.34,35 However, the detailed molecular mechanisms are implicated. Here, we suggested that circ-ZNF609 and ELF2 expression was increased and miR-188 expression was decreased in NPC. In addition, circ-ZNF609 knockdown suppressed cell proliferation and cell cycle transition, as well as induced apoptosis in NPC cells. Furthermore, circ-ZNF609 regulated NPC cell growth by modulating miR-188/ELF2axis. These findings provide potential targets for the treatment of NPC patients.
Nowadays, circRNAs are studied as a group of emerging objects in human cancers. For example, circRNA 100269 was lowly expressed and inhibited the proliferation by binding to miR-630 in gastric cancer cells.36 CircRNA0058124 induced cell invasion and tumorigenesis in papillary thyroid cancer.37 CircRNA 0001730 positively regulated cell proliferation and invasion via affecting miR-326 and Wnt7B in glioma.38 In this research, we suggested that circ-ZNF609 expression was significantly increased in NPC tissues and cells, and circ-ZNF609 depletion repressed the proliferation and cell cycle, as well as accelerated apoptosis of NPC cells. These results were consistent with the previous reporter that demonstrated that circ-ZNF609 was upregulated and promoted cell growth in NPC.15 Moreover, circ-ZNF609 was reported as a positive regulator in other human cancers, including rhabdomyosarcoma,39 renal carcinoma,40 and breast cancer.14 Therefore, circ-ZNF609 is important for NPC development.
Present evidence demonstrate that circular RNA is identified as a miRNA sponge to regulate activities of miRNAs.41 For example, circRNA ciRS-7 downregulated miR-7 expression through interaction.42 Then, we used starbase to predict the potential targets of circ-ZNF609, and found that miR-188 likely interacted with circ-ZNF609. Subsequently, this interaction was verified by the dual luciferase activity assay and RIP assay. Meanwhile, we demonstrated that circ-ZNF609 downregulated miR-188 expression in NPC cells. As a miRNA, miR-188 was reported to exert function in many types of cancer, such as colorectal cancer,43 oral squamous cell carcinoma,44 gastric cancer,45 and breast cancer.46 Besides, miR-188 acted as a regulator to repress proliferation, cycle transition, and colony formation of NPC cells.25 Agreement with this result, our data revealed that miR-188 repressed cell growth in NPC. Furthermore, circ-ZNF609 knockdown negatively regulated NPC cell growth through upregulating miR-188 expression. Although we confirmed the inhibitory function of miR-188 in NPC cells, more studies are needed for the understanding of miR-188 mechanism.
In human cancers, miRNAs are considered as mRNA sponges to mediate gene expression by binding to 3ʹ untranslated region (UTR) of downstream genes.47 Using website microT-CDS, we found that ELF2likely bound to miR-188, and then confirmed this interaction using the dual luciferase reporter assay and RIP assay. Moreover, the negative effect of miR-188 on ELF2 expression was observed in NPC cells in this assay. As a transcription, ELF2 may play a crucial role in cell development. However, the studies of ELF2 in human cancers containing NPC are less. But a paper reported that ELF2 expression was increased and ELF2 promoted cell proliferation in NPC in 2017.33 Accordance with these data, ELF2 level was upregulated in NPC cells in our assay and ELF2 positively regulated cell proliferation and cell cycle transition, and inhibited apoptosis. Moreover, miR-188 inhibited cell growth through modulating ELF2 expression. Besides, we confirmed that circ-ZNF609 downregulated miR-188 expression to increase ELF2 level in NPC cells.
In conclusion, we demonstrated that circ-ZNF609 regulated NPC cell progression via modulating miR-188/ELF2 axis, providing a theoretical basis for the treatment of NPC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. doi:10.1016/S0140-6736(15)00055-0
2. Yoshida EJ, Luu M, David JM, et al. Facility volume and survival in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2018;100(2):408–417. doi:10.1016/j.ijrobp.2017.09.038
3. Li LN, Xiao T, Yi HM, et al. MiR-125b increases nasopharyngeal carcinoma radioresistance by targeting A20/NF-kappaB signaling pathway. Mol Cancer Ther. 2017;16(10):2094–2106. doi:10.1158/1535-7163.MCT-17-0385
4. Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
5. Li LJ, Leng RX, Fan YG, et al. Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res. 2017;361(1):1–8. doi:10.1016/j.yexcr.2017.10.010
6. Spence T, Bruce J, Yip KW, et al. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):17. doi:10.21037/cco.2016.03.09
7. He R, Hu Z, Wang Q, et al. The role of long non-coding RNAs in nasopharyngeal carcinoma: as systemic review. Oncotarget. 2017;8(9):16075–16083. doi:10.18632/oncotarget.14211
8. He B, Zeng J, Chao W, et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8(25):41166–41177. doi:10.18632/oncotarget.17083
9. Lv Y, Yang H, Ma X, et al. Strand-specific miR-28-3p and miR-28-5p have differential effects on nasopharyngeal cancer cells proliferation, apoptosis, migration and invasion. Cancer Cell Int. 2019;19:187. doi:10.1186/s12935-019-0915-x
10. Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi:10.1016/j.tig.2016.03.002
11. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi:10.1080/15476286.2015.1122162
12. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi:10.1158/0008-5472.CAN-16-1883
13. Wu L, Xia J, Yang J, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J BUON. 2018;23(5):1343–1349.
14. Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res. 2018;10:3881–3890. doi:10.2147/CMAR
15. Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23(7):2817–2826. doi:10.26355/eurrev_201904_17558
16. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
17. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
18. Su Z, Yang Z, Xu Y, et al. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6(11):8474–8490. doi:10.18632/oncotarget.3523
19. Chen Z, Wang Z, Zheng C, et al. MiR-26b inhibits proliferation, invasion, and migration of glioma by targeting cyclooxygenase-2. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2017;42(2):139–146. doi:10.11817/j.issn.1672-7347.2017.02.004
20. Li H, Yang BB. MicroRNA-in drug resistance. Oncoscience. 2014;1(1):3–4. doi:10.18632/oncoscience.2
21. Friedman RC, Farh KK-H, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi:10.1101/gr.082701.108
22. Lagos-Quintana M, Rauhut R, Meyer J, et al. New microRNAs from mouse and human. Rna. 2003;9(2):175–179. doi:10.1261/rna.2146903
23. Li N, Shi H, Zhang L, et al. miR-188 inhibits glioma cell proliferation and cell cycle progression through targeting beta-catenin. Oncol Res. 2018;26(5):785–794. doi:10.3727/096504017X15127309628257
24. Zheng Y, Liu H, Kong Y. miR-188 promotes senescence of lineage-negative bone marrow cells by targeting MAP3K3 expression. FEBS Lett. 2017;591(15):2290–2298. doi:10.1002/1873-3468.12720
25. Wu J, Lv Q, He J, et al. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal. 2014;12:66. doi:10.1186/1478-811X-12-2
26. Wang CY, Petryniak B, Thompson CB, et al. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science. 1993;260(5112):1330–1335. doi:10.1126/science.8493578
27. Oettgen P, Akbarali Y, Boltax J, et al. Characterization of NERF, a novel transcription factor related to the Ets factor ELF-1. Mol Cell Biol. 1996;16(9):5091–5106. doi:10.1128/MCB.16.9.5091
28. Serdobova I, Pla M, Reichenbach P, et al. Elf-1 contributes to the function of the complex interleukin (IL)-2-responsive enhancer in the mouse IL-2 receptor alpha gene. J Exp Med. 1997;185(7):1211–1221. doi:10.1084/jem.185.7.1211
29. Sarafova S, Siu G. A potential role for Elf-1 in CD4 promoter function. J Biol Chem. 1999;274(23):16126–16134. doi:10.1074/jbc.274.23.16126
30. Lacorazza HD, Yamada T, Liu Y, et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9(3):175–187. doi:10.1016/j.ccr.2006.02.017
31. Zhang J, Hou W, Jia J, et al. MiR-409-3p regulates cell proliferation and tumor growth by targeting E74-like factor 2 in osteosarcoma. FEBS Open Bio. 2017;7(3):348–357. doi:10.1002/2211-5463.12177
32. Jin SQ, Pu YF, Qiu Y, et al. Correlation of PBX2/ELF2 expression with prognosis of non-small cell lung cancer. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2014;43(4):413–419.
33. Liu Y, Tao Z, Qu J, et al. Long non-coding RNA PCAT7 regulates ELF2 signaling through inhibition of miR-134-5p in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;491(2):374–381. doi:10.1016/j.bbrc.2017.07.093
34. Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi:10.18632/oncotarget.v8i42
35. Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. doi:10.1186/s12967-018-1582-8
36. Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9(6):1585–1594. doi:10.18632/aging.v9i6
37. Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 2019;38(1):318.
38. Lu Y, Deng X. circ_0001730 promotes proliferation and invasion via the miR-326/Wnt7B axis in glioma cells. Epigenomics. 2019. doi:10.2217/epi-2019-0121
39. Rossi F, Legnini I, Megiorni F, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38(20):3843–3854. doi:10.1038/s41388-019-0699-4
40. Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654. doi:10.1002/jcp.27744
41. Hsiao KY, Sun HS, Tsai SJ. Circular RNA – new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141. doi:10.1177/1535370217708978
42. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi:10.1158/0008-5472.CAN-13-1568
43. Yan S, Yue Y, Wang J, et al. LINC00668 promotes tumorigenesis and progression through sponging miR-188-5p and regulating USP47 in colorectal cancer. Eur J Pharmacol. 2019;858:172464. doi:10.1016/j.ejphar.2019.172464
44. Wang L, Liu H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumour Biol. 2016;37(3):4105–4113. doi:10.1007/s13277-015-4246-9
45. Li Y, Yan X, Shi J, et al. Aberrantly expressed miR-188-5p promotes gastric cancer metastasis by activating Wnt/beta-catenin signaling. BMC Cancer. 2019;19(1):505. doi:10.1186/s12885-019-5731-0
46. Pei J, Zhang J, Yang X, et al. TMED3 promotes cell proliferation and motility in breast cancer and is negatively modulated by miR-188-3p. Cancer Cell Int. 2019;19:75.
47. Ni WJ, Leng XM. miRNA-dependent activation of mRNA translation. Microrna. 2016;5(2):83–86. doi:10.2174/2211536605666160825151201
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.