Back to Journals » Cancer Management and Research » Volume 13
circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3: Selected Key circRNAs in Neuroblastoma and Their Associations with Clinical Features
Authors Lin W, Wang Z, Wang J, Yan H, Han Q, Yao W, Li K
Received 3 February 2021
Accepted for publication 7 May 2021
Published 28 May 2021 Volume 2021:13 Pages 4271—4281
DOI https://doi.org/10.2147/CMAR.S297316
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Seema Singh
Weihong Lin,* Zuopeng Wang,* Jing Wang, Hanlei Yan, Qilei Han, Wei Yao, Kai Li
Department of Pediatric Surgery, Children’s Hospital of Fudan University, National Children’s Medical Center, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Kai Li
Department of Pediatric Surgery, Children’s Hospital of Fudan University, National Children’s Medical Center, 399 Wan yuan Road, Shanghai, 201102, People’s Republic of China
Tel +86 21 64931212
Fax +86 21 64931211
Email [email protected]
Objective: The roles of circRNAs in neuroblastoma (NB) are unclear. We used next-generation sequencing to detect the circRNA expression profiles in NB to identify the key circRNAs and analyzed the relationships between the circRNAs and clinical features.
Methods: Five paired neuroblastoma tumor and adjacent normal fetal adrenal medulla samples were collected for high-throughput RNA sequencing. Bioinformatics analysis was performed for functional annotation of the host genes of differentially expressed circRNAs. Validation of dysregulated circRNAs was performed by real-time quantitative reverse transcription polymerase chain reaction. The relationships between the key circRNAs and clinical features were analyzed. In addition, overexpression of key circRNAs in an NB cell line, as well as cell proliferation assays, colony formation assays and cell migration assays, was conducted to investigate the biological functions of key circRNAs.
Results: A total of 4704 differentially expressed circRNAs were found, including 2462 up-regulated and 2242 down-regulated circRNAs. According to our previous studies, the predicted target circRNAs of miR-21 involved in tumorigenic signaling pathways were selected, including circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3. These circRNAs were associated with clinical features, and the circRNA expression was significantly lower (P < 0.05) in the NB tissues than in normal adrenal tissues. Overexpression of circRNA-TBC1D4 promotes NB cell migration, but not proliferation and colony-formation in vitro.
Conclusion: We suggest that circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3 may be cancer suppressor genes, which act by sponging miR-21 in NB. Further investigations are needed to elucidate the underlying mechanism.
Keywords: neuroblastoma, circRNA, circRNA-TBC1D4, miR-21
Introduction
Neuroblastoma (NB) is the most common extracranial malignant solid tumor in children and accounts for about 15% of cancer-associated mortality in childhood.1 The outcome remains poor due to distant metastasis at initial diagnosis.2 The prognosis for NB patients in advanced stage has not improved. Thus, exploiting effective drug targets and better biomarkers for advanced NB is of great necessity.
The pathogenic mechanism of NB remains unclear. A recent study showed that non-coding RNAs (ncRNAs) act as key regulators of physiological processes in the occurrence and development of diseases.3 Particularly relevant in cancer, ncRNAs have been identified as tumor suppressors and oncogenic drivers in diverse cancer types. NcRNAs are functional RNA species that include microRNAs (miRNAs), a class of short ncRNA (∼21–25 nucleotides), long ncRNAs (lncRNAs) consisting of more than 200 nucleotides, and circular RNAs (circRNAs) characterized by covalently closed-loop structures. They regulate gene expression post-transcriptionally and participate in diverse physiological programs such as apoptosis, autophagy, proliferation and angiogenesis.4,5 We identified hsa-miR-21 (miR-21) as a target gene of tumorigenesis in NB6 and found that LINC01296 was the most upregulated lncRNA by lncRNA microarray in advanced-stage NB tumors which was associated with poor prognosis.7
Recently, circRNAs have been reported to play a critical role in the pathogenesis of several cancers, such as ovarian cancer, hepatoblastoma and bladder cancer.8–10 The aim of this study was to investigate and validate potential therapeutic targets of circRNAs in NB.
Methods
Patients and Specimens
Tumor samples of NB and adjacent normal adrenal medulla were collected from 20 patients undergoing surgery from September 2014 to January 2019 in Children’s Hospital of Fudan University (Shanghai, China). This study was approved by the Institute Research Ethics Committee of the hospital and written informed consent was obtained from every patient. All patients were pathologically diagnosed with primary NB without preoperative chemotherapy and the samples were immediately stored at −80°C until use. Among the samples, five pairs were randomly selected for high-throughput RNA sequencing (RNA-seq) to identify differentially expressed circRNAs. The clinical information for five NB samples used is described in Table 1. We selected several of the highly upregulated and downregulated circRNAs, as well as miR-21-related circRNAs, for further analysis, and all 20 paired specimens were used for further expression validation by real-time quantitative reverse transcription polymerase chain reaction (real-time qRT-PCR). The clinical features of patients, including age, INSS stage, risk group, histologic classification, MYCN status, concentration of lactate dehydrogenase (LDH), neuron-specific enolase (NSE) and serum ferritin (SF), were also collected for further statistical analysis.
 |
Table 1 Clinical Information for Five Paired Neuroblastoma Samples Used for circRNA Sequencing |
circRNA Sequencing
Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). After using the ribo-zero kit to digest ribosomal RNA and RNase R to remove linear RNA, the enriched circRNAs were reverse-transcribed into cDNA with random hexamer primers, and then used for library construction and RNA-seq via the Illumina sequencer (Oebiotech, Shanghai, China). The raw data were processed by Trimmomatic software (http://www.usadellab.org/cms/index.php?page=trimmomatic)11 to produce high quality clean data which were then aligned to circBase12 to predict the circRNA sequences by CIRI software (https://sourceforge.net/projects/ciri/).13 Spliced reads per millions, which is the ratio of number of back-spliced junction reads to number of total reads, was utilized to quantitate the expression level of each circRNA in each sample. Correlation tests, principal component analysis (PCA) and hierarchical clustering analysis were used to show the degree of similarity of circRNA expression patterns among different specimens.
Bioinformatics Analysis
Junction read counts of each circRNA in every sample were standardized to base mean to estimate the expression level via DESeq software (in R, enter citation [“DESeq”; http://www.R-project.org]).14,15 Two criteria were applied to estimate the discrepancy of circRNA expression level between any two groups of samples: FoldChange, the multiple of change of the specific circRNA expression level in tumor compared with normal tissue, and P value, generated via negative binomial distribution test. The screening criteria set for up- and downregulated circRNAs were fold change >2.0 and a P value ≤ 0.05. Hierarchical clustering analysis was performed to display the correlation of dysregulated circRNA expression profiles among different samples. Gene Ontology (GO) analysis (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/) were performed for functional annotation of the host genes of differentially expressed circRNAs.
Validation of Dysregulated circRNAs by Real-Time qRT-PCR
Total RNA was extracted using TRIzol reagent and RNA concentration was measured by NanoDrop photospectrometer (Thermo Scientific, Waltham, MA, USA). Total RNA (1000 ng) from each sample was added into the reverse transcription system to synthesize cDNA using the PrimeScript RT reagent Kit (TaKaRa, Beijing, China). Specific primers spanning the back-splice junction site of circRNAs were designed and then synthesized (Table S1). The qRT-PCR analyses were performed using a LightCycler® 480 System (Roche, Rotkreuz, CH) with TB GREEN Premix Ex Taq II (TaKaRa). Relative RNA expression level was calculated by the 2−ΔΔCt method with GAPDH serving as internal control.
Cell Culture and circRNA-TBC1D4 Overexpression
SH-SY5Y cells were purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). The neuroblastoma cell line SH-SY5Y was cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 (DMEM/F12) (Meilunbio, Dalian, China) supplemented with 10% fetal bovine serum (FBS) (Gibco) in a humidified incubator with 5% CO2 at 37 °C. Lentiviruses LV18-circRNA-TBC1D4 and LV18NC were purchased from GenePharma (Shanghai, China). SH-SY5Y was infected with LV18-circRNA-TBC1D4, according to the manufacturer’s instructions, to overexpress circRNA-TBC1D4, and the cells were denoted SH-SY5Y-circRNA-TBC1D4. SH-SY5Y was also infected with LV18NC to serve as a control, and the cells were denoted SH-SY5Y-NC.
Cell Proliferation Assays
Cell proliferation assays were performed with Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan). Approximately 5×103 cells with 100 μL DMEM/F12 were seeded in 96-well plates. At 0, 24, 48, 72, 96 and 120 h, 10 μL CCK-8 reagent was added to each well and incubated at 37 °C for 2 h. The optical density (OD) at 450 nm was read. Every 24 h, five replicates were examined for SH-SY5Y-circRNA-TBC1D4 and SH-SY5Y-NC.
Colony Formation Assays
Approximately 2×103 SH-SY5Y-circRNA-TBC1D4 and SH-SY5Y-NC cells were seeded into six-well plates and incubated for approximately 2 weeks. Then, the colonies were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 15 min at room temperature. The cell colonies were photographed.
Cell Migration Assays
Cell migration assays were performed with Transwell permeable supports with 6.5-mm insert, 24-well plate and 8.0-μm polycarbonate membrane (Corning, NY, USA). Approximately 1×104 SH-SY5Y-circRNA-TBC1D4 and SH-SY5Y-NC cells in 100 μL DMEM/F12 without FBS were placed into the upper well, and 600 μL DMEM/F12 with 10% FBS was placed into the lower well. After incubation for 36 h, the cells on the inserts were fixed with 4% paraformaldehyde for 30 min, stained with 1% crystal violet for 15 min and then photographed.
Statistical Analysis
Statistical analysis was performed by SPSS 22.0 (IBM, Armonk, NY, USA). Graphical depiction of data was generated by GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA). Comparison of circRNA expression levels between NB and adjacent normal fetal adrenal medulla was analyzed by paired Student t-test. Correlations between dysregulated FoldChange of specific circRNAs and other clinical data were performed by Spearman correlation analysis. A P value <0.05 was considered statistically significant.
Results
Expression Profiles of circRNAs in NB (Reference PRJNA721263 in SRA Database)
A total of 57,857 circRNAs were identified in five randomly selected pairs of samples by RNA-seq. The number of identified circRNAs in each sample is shown in Figure S1A. The average length was 3315.65 bp, range 38–99,918 bp (detailed length distribution is shown in Figure S1B). GC content frequency distribution and chromosome/scaffold distribution were also analyzed (Figure S1C and D). The most common circRNA form analyzed by FEELnc software16 was sense (direction), genetic (type) and exonic (location), which accounted for 90.7% of total circRNA (Figure S1E). Correlation tests, PCA, as well as hierarchical clustering analysis, all suggested that different expression patterns of circRNAs could be distinguished in the NB group and adjacent normal fetal adrenal medulla control group, but expression patterns were very similar in the same group (Figure S1F, G and H).
Bioinformatics Analysis of Differentially Expressed circRNAs
With the screening criteria for up- and downregulated circRNAs set as FoldChange >2.0 and P < 0.05, 4704 differentially expressed circRNAs were identified between NB and adjacent normal fetal adrenal medulla, including 2462 upregulated and 2242 downregulated circRNAs. Volcano plots visualized the significant differentially expressed circRNAs between the two groups (Figure 1A). The unsupervised hierarchical clustering analysis of differentially expressed circRNAs showed that the expression patterns of dysregulated circRNAs were distinguishable between the NB group and adjacent normal fetal adrenal medulla group (Figure 1B). Functional annotation of the host genes of differentially expressed circRNAs was conducted through GO analysis and KEGG pathway analysis. The most significantly enriched GO terms in biological process, cellular component and molecular function were mRNA pseudouridine synthesis, plasma membrane and acetyl-CoA carboxylase activity, respectively (the top 10 significantly enriched GO terms in the above three categories are shown in Figure 1C). KEGG pathway analysis showed that the most related pathways were fluid sheer stress and atherosclerosis (Figure 1D).
Validation of Selected Differentially Expressed circRNA by Real-Time qRT-PCR
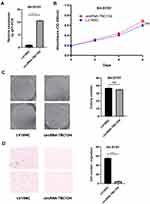
Among the 4704 differentially expressed circRNAs, 10 highly upregulated circRNAs and five highly downregulated circRNAs, which are associated with tumorigenesis and progression, were selected for expression validation by real-time qRT-PCR. Eight of 10 upregulated circRNAs and two of five downregulated circRNAs were finally verified in 20 paired samples (Figure 2A and B). Our previous study had shown that miR-21 plays a significant role in the regulation of proliferation and apoptosis in NB,6,17 so we selected another seven circRNAs for analysis, whose predicted target miRNA was miR-21. Real-time qRT-PCR confirmed that three miR-21-related circRNAs, including circRNA-TBC1D4 (average FoldChange = 18.7; P = 0.0002), circRNA-NAALAD2 (average FoldChange = 5.6; P = 0.0032) and circRNA-TGFBR3 (average FoldChange = 23.3; P = 0.0079), were significantly downregulated in NB samples (Figure 2C). The binding sites are shown in Figure 3. Detailed information about the 22 selected circRNAs are described in Table S1.
 |
Figure 3 Binding sites between miR-21 and (A) circRNA-TBC1D4, (B) circRNA-NAALAD2 and (C) circRNA-TGFBR3. |
Correlation Between Significantly Downregulated miR-21-Related circRNAs and Clinical Features
To determine whether circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3 were associated with clinical characteristics of NB, Spearman correlation analysis was performed to calculate the correlation coefficients between downregulated FoldChange of circRNA-TBC1D4, circRNA-NAALAD2, circRNA-TGFBR3 and clinical features in 20 patients (Table 2). The results showed that circRNA-TBC1D4 was associated with MYCN status (P = 0.0488; R = 0.6344) and the concentration of LDH (P = 0.0109; R = 0.7590), circRNA-NAALAD2 was associated with the concentration of LDH (P = 0.0214; R = 0.7101), and circRNA-TGFBR3 was associated with the histology category (P = 0.0001; R = 0.9692). In addition, we also found that downregulated circRNA-TBC1D4, circRNA-NAALAD2, circRNA-TGFBR3 were associated with each other, indicating that the three circRNAs may be downregulated simultaneously in NB (Table 2).
 |
Table 2 Correlations of Fold Decreases Among circRNA-TBC1D4, circRNA-NAALAD2, circRNA-TGFBR3 and Clinical Features in 20 Patients |
Overexpression of circRNA-TBC1D4 Promotes CRC Cell Migration
In order to investigate the biological functions of circRNA-TBC1D4 in neuroblastoma, we overexpressed circRNATBC1D4 in the SH-SY5Y neuroblastoma cell lines, because circRNA-TBC1D4 was downregulated in neuroblastoma samples. LV18-circRNA-TBC1D4 infection significantly increased circRNA-TBC1D4 expression in SH-SY5Y cells (Figure 4A). Cell proliferation assays using CCK8 showed that overexpression of circRNA-TBC1D4 did not decrease the proliferation of SH-SY5Y cells (Figure 4B). Colony formation assays showed that overexpression of circNRA-TBC1D4 did not decrease the colony-forming ability of SH-SY5Y cells (Figure 4C). However, cell migration assays showed that overexpression of circNRA-TBC1D4 significantly decreased the migration ability of SH-SY5Y cells (Figure 4D).
Discussion
The circRNAs belong to the family of ncRNAs implicated in cancer progression and tumorigenesis. Emerging studies have shown that circRNAs are ubiquitous in eukaryotes and play essential roles in modulating gene expression and signaling pathways by sponging miRNA or other molecules including proteins that participate in carcinogenesis.18 Some studies have found that circRNA can be used as a diagnostic or prognostic indicator. For instance, Peng et al reported that hsa_circ_0081001 may act as a diagnostic biomarker in osteosarcoma because it is upregulated in tissues and serums and is correlated the poor prognosis.19 Chen et al suggested that hsa_circ_0000190 may have better sensitivity and specificity in the diagnosis of gastric cancer compared with commonly used biomarkers such as CEA and CA19-920 Other studies have found that circRNA can be a potential therapeutic target which is associated with cancer pathogenesis. Liu et al found that circ0001361 plays an oncogenic role in bladder cancer invasion and metastasis by targeting the miR-491-5p/MMP9 axis, and this circRNA may be a potential novel target for bladder cancer therapy.21 Hsa_circ_0001944 is another tumor-promoting circRNA in bladder cancer that functions as an endogenous miRNA competitor to regulate PROK2 expression by binding to miR-548.8 Chen et al suggested that oncogenic circAGO2 drives cancer progression by facilitating HuR-repressed functions of AGO2-miRNA complexes. Hsa_circ_000984 affects colorectal cancer cell invasion, migration and growth by competing with cell cycle-associated proteins for sponging of miR-106b.22
Despite the above studies, the expression and function of circRNAs in NB remain unclear. In the present study, we screened circRNA expression profiles in five paired NB samples by RNA sequencing. The results indicated there were 2462 upregulated and 2242 downregulated circRNAs. Our previous study focused on the oncogenic gene miR-21 in NB and found that miR-21 may regulate the potential targets PTEN/PDCD4 to promote apoptosis.6,17 In addition, miR-21 functions as a significant common carcinogen in a variety of cancers, such as gliomas,23 colon cancer,24 cutaneous malignant melanoma25 and retinoblastoma.26 Three miR-21-related circRNAs, circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3, were confirmed to be overexpressed in adjacent adrenal tissue compared to tumor tissue and this was verified by qRT-PCR. Furthermore, we correlated these circRNAs with clinical features, which indicated that circRNA-TBC1D4 was associated with MYCN status and value of LDH, while circRNA-NAALAD2 was associated with value of LDH and circRNA-TGFBR3 was associated with the Favorable Histology category. These correlations showed that patients with NB with downregulation of circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3 expression might have poorer clinical outcomes. Meanwhile, circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3 were correlated with each other, indicating that the three circRNAs may synergistically downregulate the expression of miR-21 in NB. We additionally overexpressed circRNA-TBC1D4 in the NB cell line SH-SY5Y. Cell proliferation assays, colony formation assays and cell migration assays showed that overexpression of circRNA-TBC1D4 promotes NB cell migration, but not proliferation and colony-formation in vitro. These results suggested that circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3 are often downregulated simultaneously in NB, and may serve as biomarkers indicating an unfavorable NB type.
Conclusion
In conclusion, we performed high-throughput sequencing of NB. Through clinical correlation analysis and bioinformatics analysis, we determined that circRNA-TBC1D4, circRNA-NAALAD2 and circRNA-TGFBR3 were cancer suppressor genes, which act by sponging miR-21. We intend in future studies to further investigate the effects of these genes on the biological behavior of NB in vivo and in vitro.
Ethics Approval and Consent to Participate
I confirm that I have read the Editorial Policy pages. This study was conducted with approval from the Ethics Committee of the Children’s Hospital of Fudan University. This study was conducted in accordance with the declaration of Helsinki. Written informed consent was obtained from all patients or guardians enrolled in this study.
Consent for Publication
All participants signed a document of informed consent.
Acknowledgments
We would like to acknowledge the hard and dedicated work of all the staff that implemented the intervention and evaluation components of the study. Weihong Lin and Zuopeng Wang are co-first authors for this study.
Disclosure
The authors declare that they have no competing interests.
References
1. Maris JM, Hogarty MD, Bagatell R, et al. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. doi:10.1016/S0140-6736(07)60983-0.
2. Ma Y, Zheng J, Feng J, et al. Neuroblastomas in Eastern China: a retrospective series study of 275 cases in a regional center. PeerJ. 2018;6:e5665. doi:10.7717/peerj.5665.
3. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi:10.15252/embj.2018100836.
4. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2020;21:22–36. doi:10.1038/s41568-020-00306-0.
5. Cannavicci A, Zhang Q, Kutryk M. Non-Coding RNAs and hereditary hemorrhagic telangiectasia. J Clin Med. 2020;9(10):3333. doi:10.3390/jcm9103333.
6. Wang Z, Yao W, Li K, et al. Identification of hsa-miR-21 as a target gene of tumorigenesis in neuroblastoma. Int J Clin Exp Med. 2016;9(2):2718–2727.
7. Wang J, Wang Z, Yao W, et al. The Association between lncRNA LINC01296 and the Clinical Characteristics in Neuroblastoma. J Pediatr Surg. 2019;54(12):2589–2594. doi:10.1016/j.jpedsurg.2019.08.032.
8. Jin M, Lu S, Wu Y, et al. Hsa_circ_0001944 promotes the growth and metastasis in bladder cancer cells by acting as a competitive endogenous RNA for miR-548. J Exp Clin Cancer Res. 2020;39(1):186. doi:10.1186/s13046-020-01697-6.
9. Liu BH, Zhang BB, Liu XQ, et al. Expression profiling identifies circular RNA signature in hepatoblastoma. Cell Physiol Biochem. 2018;45(2):706–719. doi:10.1159/000487163.
10. Li L, Yu P, Zhang P, et al. Upregulation of hsa_circ_0007874 suppresses the progression of ovarian cancer by regulating the miR‐760/SOCS3 pathway. Cancer Med. 2020. doi:10.1002/cam4.2866.
11. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
12. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi:10.1261/rna.043687.113.
13. Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16(1):4. doi:10.1186/s13059-014-0571-3.
14. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi:10.1186/gb-2010-11-10-r106.
15. Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008–2017. doi:10.1101/gr.133744.111.
16. Wucher V, Legeai F, Hédan B, et al. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017;45(8):e57. doi:10.1093/nar/gkw1306.
17. Wang Z, Yao W, Li K, et al. Reduction of miR-21 induces SK-N-SH cell apoptosis and inhibits proliferation via PTEN/PDCD4. Oncol Lett. 2017;13(6):4727–4733. doi:10.3892/ol.2017.6052.
18. Cortés-López M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89(4):527–537.
19. Kun-Peng Z, Chun-Lin Z, Jian-Ping H, et al. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14(11):1513–1520. doi:10.7150/ijbs.27523.
20. Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clinica Chimica Acta. 2017;466:167–171. doi:10.1016/j.cca.2017.01.025.
21. Liu F, Zhang H, Xie F, et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2019. doi:10.1038/s41388-019-1092-z.
22. Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683. doi:10.18632/oncotarget.21748.
23. Kwak HJ, Kim YJ, Chun KR, et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 2011;30(21):2433–2442. doi:10.1038/onc.2010.620.
24. Liu M, Tang Q, Qiu M, et al. miR-21 targets the tumor suppressor RhoB and regulates proliferation, invasion and apoptosis in colorectal cancer cells. FEBS Lett. 2011;585(19):2998–3005. doi:10.1016/j.febslet.2011.08.014.
25. Jiang L, Lv X, Li J, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114(6):582–588. doi:10.1016/j.acthis.2011.11.001.
26. Shen F, M H M, Chen L, et al. MicroRNA-21 Down-regulates Rb1 expression by targeting PDCD4 in retinoblastoma. J Cancer. 2014;5(9):804–812. doi:10.7150/jca.10456.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.