Back to Journals » Cancer Management and Research » Volume 12
circRNA MYLK Accelerates Cervical Cancer via Up-Regulation of RHEB and Activation of mTOR Signaling
Authors Chen R, Mao L, Shi R, Wang W, Cheng J
Received 11 November 2019
Accepted for publication 18 March 2020
Published 19 May 2020 Volume 2020:12 Pages 3611—3621
DOI https://doi.org/10.2147/CMAR.S238172
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Rui Chen,1 Luning Mao,2 Rui Shi,1 Wenjing Wang,1 Jingxin Cheng1
1Department of Gynecology, East Hospital Affiliated to Tongji University, Shanghai 200012, People’s Republic of China; 2Department of Pathology, Basic Medical College, Fudan University, Shanghai 200032, People’s Republic of China
Correspondence: Jingxin Cheng
Department of Gynecology, East Hospital Affiliated to Tongji University, No.1800 Yuntai Road, Pudong New Area, Shanghai 200012, People’s Republic of China
Email [email protected]
Background: Growing evidence directly suggested that circular RNAs (circRNAs) are crucial contributors in the course of cervical cancer (CC) onset and progression. Nevertheless, a large number of circRNAs have not been fully addressed in their function and underlying mechanisms during CC etiology.
Purpose: Our study focused on the function of circRNA MYLK (myosin light chain kinase), one novel tumor-related circRNA, in CC cell behaviors.
Methods: Firstly, we evaluated the expression profile of circMYLK in CC cells and in normal Ect1/E6E7 cell line. Moreover, the accurate function of circMYLK in CC cells was assessed via colony formation, CCK-8, EdU, and TUNEL assay. The association among circRNAs, miRNA, and target mRNAs was predicated by bioinformatics methods and validated in mechanical assays.
Results: We disclosed that circMYLK was up-regulated in CC cell lines and acted as a sponge of miR-1301-3p. Besides, downstream miR-1301-3p was capable of reversing circMYLK-mediated CC cell growth and apoptosis. Furthermore, we validated that circMYLK bound to miR-1301-3p as a sponge to upregulate RHEB (Ras homolog, mTORC1 binding) expression. As annotated in prior works, RHEB was responsible for mTOR signaling transduction. Therefore, we investigated whether circMYLK functioned its tumor-facilitating impact in CC through a RHEB-dependent mTOR signaling activation.
Conclusion: It was unveiled that circMYLK sponged miR-1301-3p to promote RHEB expression, which resulted in mTOR signaling activation and CC cell malignant growth.
Keywords: circMYLK, miR-1301-3p, RHEB, mTOR signaling, cervical cancer
Introduction
Cervical cancer (CC) has become a public health threat among females, ranking the fourth among the most commonly occurred tumors. Overall, there are about 528,000 new cases of CC in 2012.1 Globally, CC-induced mortalities in 2012 are approximately 266,000, taking up 7.5% of all female cancer deaths. It is estimated that by the year of 2030, this number will climb to 410,000.2 Therefore, it is of great significance to deeply investigate the underlying mechanism about CC etiology.
As annotated before, the activation of cervical cancer is strongly related with non-coding RNAs. In tumor biology, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) recognized as two main components of non-coding RNAs (ncRNAs), are widely addressed owing to their great contributions.3–5 As newly emerged ncRNAs, circular RNAs (circRNAs) are also essentially involved in tumor formation and progression.6,7 Forty-eight years ago, circRNAs existence was uncovered. However, circRNAs were not thoroughly understood and were regarded as incorrect gene rearrangements or splicing mistakes.8 Owing to high-throughput sequencing, numerous circRNAs have been functionally analyzed. Basically, circRNAs are largely exonic circRNAs derived from parental gene exons.9,10 Exonic circRNAs are covalently heat-to-tail closed and structured in a loop without 5′ end or a 3′ end, resulting in higher resistance and stability to RNA exonuclease.11,12
Additionally, the crucial functions of circRNAs in tumorigenesis include miRNA sponges,13 protein sponges14,15 and translation contributors.16 Basically, the most reported function of circRNAs is the sponge-like property in tumors. Numerous mRNAs or circRNAs share binding sites with miRNAs and a competition between mRNAs or circRNAs to interact with miRNAs is formed in regulating cancer development, which is called the pattern of competing endogenous RNA (ceRNA).17 For example, the “miRNA sponge” role of hsa_circ_0007534 as a miR-498 sponge to regulate BMI-1 is certified in CC cellular proliferation and invasion.18,19
mTOR is extensively corroborated as a crucial downstream molecule of AKT1. As one classic signaling pathway, the AKT/mTOR pathway mediates the metabolic homeostasis in tumor, which is conducive to uncontrolled tumor growth and metastasis.20 In gastric cancer, the AKT/mTOR axis contributes to cell proliferation, cell viability, cell cycle G1/S transition, and migration.21 mTORC1 (mechanistic target of rapamycin complex 1) is well-defined to facilitate the Warburg effect and accelerate tumor growth by sustaining the highly proliferative feature of cancer cells. The mTOR function and implication has been extensively recorded in multiple tumors such as breast cancer,22 hepatocellular carcinoma,23 and CC.24,25 Furthermore, the anti-tumor approaches have been proposed using mTOR inhibitors in CC.26,27 However, mechanism explanation about mTOR pathway is limited in CC.
CircMYLK originates from MYLK (myosin light chain kinase) and is an oncogenic factor in bladder cancer,28 prostate cancer29 and laryngeal squamous cell carcinoma.30 Our work was designed to address the function of circMYLK in CC cells. Moreover, whether circMYLK could regulate mTOR axis through a ceRNA way in CC was probed.
Materials and Methods
Cell Culture and Treatment
CC cell lines (DoTc2 4510, HCC94, C-33A, HT3) and control Ect1/E6E7 cells were applied in present study. HCC94 cell lines were purchased commercially from Cell bank of Chinese Academy of Sciences while other cell lines were purchased commercially from ATCC (American Type Culture Collection). Cells were allowed to grow in RPMI-1640 medium (Gibco, Grand Island, NY, USA) at 37oC with 5% CO2 for this study, obtained from Shanghai Institute of Cell Biology (Shanghai, China). Ten percent fetal bovine serum (FBS; Gibco), 100 μg/ml of streptomycin and 100 U/ml of penicillin were employed as medium supplements. 3 U/μg of RNase R (Ribonuclease R) from Epicentre Technologies (Madison, WI, USA) was added to culture medium for 30 min at 37oC for treating cells.
Quantitative Real-Time PCR (qRT-PCR)
Total cellular RNAs were isolated from HCC94 and C-33A cells based on the established protocol of TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNAs from each sample were employed for synthesizing cDNA (complementary DNA) which was subjected to qPCR in presence of SYBR Green kit (Takara, Tokyo, Japan). In the process of q-PCR experiments, 1μg RNA was applied. The RNA concentration taken to RT reaction was 365ng/μL, OD260/OD280=1.8. The RIN value is 10 and there were no degradation conditions. The results were calculated with 2−ΔΔCt method, with U6 and GAPDH as internal references.
Nucleic Acid Electrophoresis
The cDNA and gDNA (genomic DNA) PCR results of circMYLK were examined by agarose gels in presence of TE buffer from Thermo Scientific (Waltham, MA, USA). After electrophoresis at 110 V, DNA was separated and detected by UV irradiation.
Cell Transfection
HCC94 or C-33A cells in the 6-well plates were cultivated to 80% confluence for transfection using Lipofectamine 2000 kit (Invitrogen) for 48 h. The two circMYLK-specific shRNAs (short hairpin RNAs) (sh/circMYLK#1/2) and control shRNA (sh/ctrl), as well as pcDNA3.1(+) CircRNA Mini Vector for circMYLK, pcDNA3.1 vector for RHEB (Ras homolog, mTORC1 binding) and their relative empty vectors as control, were all purchased from Genepharm (Shanghai, China). Additionally, miR-1301-3p mimics and NC (negative control) mimics, miR-1301-3p inhibitor and NC inhibitor, were also produced by Genepharm. The relation of transfection reagent to plasmid/vector amount is 2:1. Per well in the 6-well plate, and 2.5μg plasmids were transfected. Then, 200 μmol microRNA (mimic/inhibitor and NC control) was used per well in the 6-well plate.
Colony Formation
HCC94 and C-33A cells after transfection were transplanted to the 6-well plates with the density of 1 × 103/100 μL for 14 days of incubation at 37°C with 5% CO2. After removing medium, colonies in phosphate-buffered saline (PBS) were subjected to fixation and staining by 4% paraformaldehyde and 0.1% crystal violet.
Cell Counting Kit-8 (CCK-8)
CC cells (HCC94 and C-33A) in 96-well plates at the density of 1 × 103 per well were mixed with 10 μL of CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) at indicated time points for 12 h. Absorbance was read using a microplate reader at 450 nm.
EdU (5-Ethynyl-2ʹ-Deoxyuridine) Incorporation Assay
5 × 105 HCC94 and C33A cells were seeded in 12-well plates with Cell-light™ EdU ApolloR567 In Vitro Imaging Kit (Ribobio, Guangzhou, China) for 4 h, then fixed in 4% paraformaldehyde for 30 min. After permeabilization in 0.5% TritonX-100, cell nuclei were treated with DAPI (4ʹ,6-diamidino-2-phenylindole) solution and imaged by fluorescent microscope.
TUNEL (Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling) Staining
TUNEL assay was run in the presence of In Situ Cell Death Detection Kit (Roche Diagnostics GmbH, Mannheim, Germany). The HCC94 and C33A cells in 4% paraformaldehyde were permeabilized in 100% methanol for 5 min and rehydrated. Cells were treated with fluorescein-TUNEL reagent and then with DAPI staining. Twelve-well plate was used and cell density was at 80% confluence. Apoptotic cells were visualized using inverted microscope (Olympus, Tokyo, Japan).
Subcellular Fractionation Assay
The lysed HCC94 and C33A cells in cell fractionation buffer were centrifuged for separating cell nucleus and cytoplasm. A PARIS Nuclear/cytoplasmic separation kit was used for subcellular fractionation assay. The remaining lysates were incubated with cell disruption buffer to lyse cell nuclei. The centrifugation condition was 10,000xg for 30s-1min. The relative expression levels of GAPDH as cytoplasm control, U1 as nucleus control and circMYLK were determined by qRT-PCR.
Fluorescence in situ Hybridization (FISH)
HCC94 and C33A cells in 0.5% Triton X-100 and 1 × PBS were collected and cultured with RNA FISH probe for circMYLK in hybridization solution all night at 37oC. Twelve-well plate was used and cell density was at 30% confluence. Following washing in sodium citrate buffer (SSC) in the dark, cells were counterstained with DAPI and observed under microscope (Leica, Wetzlar, Germany).
Dual-Luciferase Reporter Assay
2.5 × 105 HCC94 and C33A cells were seeded per well in a 6-well plate. Dual-luciferase reporter vectors circMYLK-WT (wild type) and RHEB-WT reporters were formed via cloning circMYLK sequence or 3ʹ-UTR (untranslated region) of RHEB, which both contained the miR-1301-3p binding sites, into pmirGLO Dual-Luciferase Vector (Promega, Madison, WI, USA). The mutant vectors circMYLK-MUT (mutant) and RHEB-MUT were designed using point mutations of miR-1301-3p binding sites. HCC94 and C-33A cells were co-transfected with reporter vectors and indicated transfection plasmids for 48 h, and finally detected by Dual-Luciferase Reporter Assay System (Promega). The relation of transfection reagent to plasmid/vector amount is 2:1 with 2.5μg transfected plasmids per well.
RNA Immunoprecipitation (RIP)
8 × 107 HCC94 and C33A cells were cultured in 10 cm cell culture dish (the number of cell dish is 7). Cells were reaped from RIP lysis buffer and immunoprecipitated with anti-Ago2 (Argonaute 2) antibody (Millipore, Bedford, MA, USA). Five microgram anti-Ago2 antibody was used in this assay with the dilution of 1:20 (v:v). Anti-IgG antibody was seen as negative control. The precipitates were purified and analyzed by qRT-PCR.
RNA Pull-Down
3 × 107 cells were planted in 10 cm cell culture dish (the number of cell dish is 3). Protein extracts from HCC94 or C-33A cells were subjected to incubation with biotinylated miR-1301-3p probes (Bio-miR-1301-3p-wt, Bio-miR-1301-3p-mut, Bio-NC) and magnetic beads. Probes concentration was 100 pmol. The fold enrichment of circMYLK in RNA-protein complex was measured by qRT-PCR.
Western Blotting
5 × 106 HCC94 or C-33A cells with 90% confluence in 6-well plates were incubated with lysis buffer and then centrifuged. Forty microgram of protein samples were electrophoresed in 10% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and transferred to PVDF (polyvinylidene fluoride) membranes. The respective bands were mixed all night with primary antibodies against RHEB (ab92313), EIF5 (eukaryotic translation activation factor-5) (ab228874), mTORC1 (ab32028) and GAPDH (ab181602), as well as relative anti-IgG secondary antibody (all from Abcam, Cambridge, MA, USA). The dilution of primary and secondary antibody is as follows: RHEB: 1/2000; EIF5: 1/1000; mTORC1: 1/1000; GAPDH: 1/10,000; Goat Anti-Rabbit IgG H&L (HRP) (ab6721): 1/5000. Enhanced chemiluminescence (ECL) detection system (Bio-Rad lab, Hercules, CA, USA) was employed for detection of signal.
Statistical Analyses
All data acquired from all three different bio-replications were analyzed by Prism 6.0 (GraphPad, San Diego, CA, USA) and presented as mean ± SD (standard deviation). The probability (p-value) between groups was assessed by Student’s paired samples t-test or ANOVA (analysis of variance), with p < 0.05 as threshold.
Results
Silencing Circular Transcript MYLK Inhibits CC Cell Growth
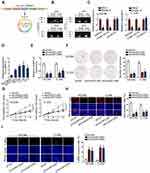
As demonstrated, circMYLK were derived from MYLK (Figure 1A). To assess the involvement of circMYLK in CC, firstly we confirmed the circular presence of circMYLK. Two sets of primers including divergent primers and convergent primers were designed for the amplification of circular MYLK or linear MYLK transcript, separately. CircMYLK was produced by divergent primers in cDNA but not gDNA, whereas linear MYLK was amplified in both cDNA and gDNA by convergent primers (Figure 1B). Additionally, RNase R experiment indicated the higher resistance of circMYLK compared to linear MYLK (Figure 1C). Next, we compared the expression of circMYLK in CC cell lines and Ect1/E6E7 (control cells). Actively expressed circMYLK was observed in CC cell lines, indicating its underlying participation in CC (Figure 1D). C-33A and HCC94 were the top two overexpressed cell lines and therefore were selected for the loss-of-function assays. Applying the back-splice junction-specific shRNAs (sh/circMYLK#1, sh/circMYLK#2), we knocked down circMYLK expression in C-33A and HCC94 cells (Figure 1E). As analyzed in cell proliferation assays, depletion of circMYLK remarkably restrained cell cloning potency, viability and proliferation ability (Figure 1F–H). Conversely, circMYLK-deficient cells presented enhanced apoptosis ability relative to control cells (Figure 1I). Collective findings corroborated that depletion of circMYLK exerted anti-proliferation impact in CC cells.
CircMYLK Sponges miR-1301-3p in CC Cells
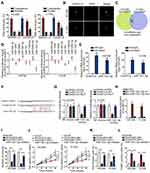
The mechanism of circMYLK was investigated. Prior to that, we assessed the cytoplasmic and nuclear distribution of circMYLK. Results revealed the major presence of circMYLK in cytoplasm (Figure 2A). Then, FISH assay re-ensured that circMYLK dominantly existed in the cytoplasm of HCC94 and C-33A (Figure 2B). Considering that cytoplasmic circRNAs may sponge miRNAs, we next searched the candidate miRNAs of circMYLK. A starBase tool (http://starbase.sysu.edu.cn/) predicted the top 50 miRNAs. Afterwards, we probed the significantly downregulated miRNAs in both HCC94 and C-33A in comparison to Ect1/E6E7 and eight miRNAs (miR-1301-3p, miR-624-3p, miR-1294, miR-449a, miR-363-3p, miR-761, miR-1306-5p, miR-526b-5p) were identified (Figure 2C). In advance of the luciferase reporter assay, overexpression efficiency of eight miRNAs was verified (Figure S1A). It turned out that only miR-1301-3p mimics declined circMYLK luciferase activity (Figure 2D). Former study pointed that Ago2 protein interacts with both miRNAs and circRNAs for the formation of RNA-induced silencing complex. Therefore, RIP experiment was utilized to validate whether circMYLK and miR-1301-3p could bind to Ago2 protein. Experimental data indicated that miR-1301-3p and circMYLK were abundantly enriched in the Ago2-precipitated RISC (RNA induced silence complex) (Figure 2E). The binding sites between wild/mutant circMYLK and miR-1301-3p were shown (Figure 2F). Results of luciferase reporter analysis demonstrated that wild circMYLK luciferase activity was decreased in response to miR-1301-3p mimics, but the luciferase activity was not altered by miR-1301-3p mimics when the putative miR-1301-3p binding sites were mutated (Figure 2G). In addition, RNA pull-down assay indicated that biotinylated wild type miR-1301-3p (Bio-miR-1301-3p-wt) pulled down circMYLK while the mutant counterpart (Bio-miR-1301-3p-mut) failed (Figure 2H). Moreover, we also functionally validated the regulation of circMYLK/miR-1301-3p on CC cell growth. As was revealed in colony formation assay, the number of colonies was declined following circMYLK downregulation while suppressing miR-1301-3p rescued this decline (Figure 2I). Furthermore, the CC cell viability and proliferation suppression caused by circMYLK downregulation could be reversed by the depletion of miR-1301-3p (Figure 2J and K). The pro-apoptosis activity of sh/circMYLK#1 was attenuated by miR-1301-3p inhibitor in CC cells (Figure 2L). Therefore, we concluded that miR-1301-3p was required for circMYLK-mediated CC development.
CircMYLK Sponges miR-1301-3p to Activate RHEB-Dependent mTOR Pathway
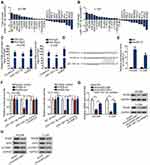
In view of post-transcriptional regulation of miRNAs via base-paring with mRNAs 3′UTR, target mRNAs of miR-1301-3p were provided by bioinformatics logarithms. Top 25 target mRNAs were demonstrated to possess highly conserved miR-1301-3p binding sequence. Afterwards, the fold changes of 25 predicted mRNAs were measured in miR-1301-3p-overexpressing cells and control cells. Specifically, RHEB expression decreased the most when miR-1301-3p was upregulated (Figure 3A and B). RIP assay described the abundant enrichment of circMYLK, RHEB and miR-1301-3p in RISC (Figure 3C). The interacting sequence of RHEB and miR-1301-3p was exhibited (Figure 3D). Mechanical assay about their interaction was conducted subsequently. CircMYLK was significantly up-regulated in HCC94 and C-33A cells (Figure 3E). Luciferase reporter assay manifested that the luciferase activity of RHEB-wt reporter was decreased by miR-1301-3p but was recovered by circMYLK overexpression. No significant alterations were found in RHEB-mut or control reporters (Figure 3F). Afterwards, we downregulated miR-1301-3p expression in CC cells (Figure S1B). CircMYLK depletion resulted in RHEB mRNA decrease, but miR-1301-3p inhibitor reversed this decrease. Similar results were also generated in western blot. The decreased protein level of RHEB induced by circMYLK depletion was increased by miR-1301-3p inhibitor (Figure 3G). As well identified, RHEB was a potent activator in mTOR signaling transduction.31,32 Above data offered us a hint that circMYLK may contribute to RHEB/mTOR signaling activation. To monitor the mTOR signaling activity in CC cells, we investigated the expression of mTOR pathway-related proteins including RHEB, EIF5, and mTORC1. Results demonstrated that circMYLK depletion led to lowly expressed level of mTOR signaling related genes (Figure 3H), indicating an impaired mTOR signaling by silenced circMYLK. Collectively, oncogenic circMYLK in CC facilitated RHEB/mTOR signaling activation via sponging miR-1301-3p to up-regulate RHEB.
The Oncogenic Activity of circMYLK in CC Depends on RHEB/mTOR Signaling Activation
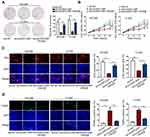
The function role of circMYLK/miR-1301-3/RHEB was elucidated in rescue experiments. RHEB expression was enforced by RHEB-overexpressing plasmids (Figure S1C). CC cell proliferation and viability was evaluated. Experimental data displayed that sh/circMYLK#1-caused proliferation suppression was countervailed by RHEB re-introduction, as elucidated in colony formation, CCK-8, and EdU (Figure 4A–C). In addition, TUNEL analysis elaborated that sh/circMYLK#1-induced apoptosis enhancement could be abolished by the upregulation of RHEB (Figure 4D). CircMYLK triggered RHEB/mTOR signaling and promoted CC cell malignant behaviors.
Discussion
Cervical cancer (CC) is gradually becoming a public health threat among females, ranking the fourth among the most commonly occurred tumors. In the fields of molecular basis of CC, wide attention has been poured into lncRNAs and protein-coding genes. Whereas circRNAs, characterized as cancer drivers, are not well elucidated in terms of their expression or potential mechanical strategies. CC-implicated circRNAs such as circ_0067934 and circ_0000745 are reported to strengthen the invasion, migration and proliferation capacities of CC cells.33,34 CircMYLK is an oncogenic factor in bladder cancer,28 prostate cancer,29 laryngeal squamous cell carcinoma30 and hepatocellular carcinoma.35 Herein, circMYLK was actively expressed in CC cells. Besides, we dissected that circMYLK also acted as a CC inducer through its anti-apoptotic and pro-proliferative properties, which further enriched the pathogenesis of CC.
It is often presented that circRNAs possess the miRNA binding sites, which is responsible for miRNA association, and restores the restrictive function of miRNA on target genes. In this study, circMYLK was widely presented in the cytoplasm of CC cells. We unmasked that miR-1301-3p was sponged by circMYLK. In the meantime, we consolidated that downregulating miR-1301-3p could counteract the growth-suppressive trend of CC cells induced by circMYLK depletion.
RHEB is identified as a conserved small GTPase and pertains to the Ras superfamily.36 The close relationship between RHEB and proliferative disorder has been revealed in multiple cancers. Primarily, RHEB is attributed to proliferation activation.37,38 Furthermore, RHEB is an essential upstream modulator of the mammalian target activity of mTOR signaling.39 Although studies demonstrated the frequent hyper-activation of mTOR signaling during CC progression, we still lack of comprehensive understanding about its participation. For example, de Melo et al. unmasked the potential usage of mTOR-targeted therapy in gynecologic cancers.25 Xie et al. elucidated that NVP-BEZ235 blocks mTOR signaling and suppresses CC growth.40 In this study, we confirmed the facilitating role of RHEB in activating mTOR pathway. On the other hand, mechanical studies demonstrated that RHEB was targeted and repressed by miR-1301-3p while circMYLK could rescue it from miR-1301-3p repression, leading to mTOR signaling activation. Therefore, we dissected that circMYLK mediated tumor-facilitating effect in CC depended on RHEB-induced mTOR signaling activation through sponging miR-1301-3p. Moreover, rescue experiments showed that RHEB re-expression abolished the suppressive impact of circMYLK silencing in CC cellular growth.
Conclusions
To conclude, our work first unveiled the high expression and oncogenic function of circMYLK in CC cells. Then, circMYLK sponged miR-1301-3p to up-regulate RHEB and resulted in mTOR signaling activation in the development of CC, shedding a new insight in future treatment of CC.
Data Sharing Statement
Data and material are not shared.
Acknowledgments
We thank for the experimental assistance from involved members. Rui Chen and Luning Mao are co-first authors for this study.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi:10.3322/caac.21166
2. Goetze OT, Al-Batran SE, Chevallay M, Monig SP. Multimodal treatment in locally advanced gastric cancer. Updates Surg. 2018;70(2):173–179. doi:10.1007/s13304-018-0539-z
3. Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16(1):107. doi:10.1186/s12943-017-0671-2
4. Zhang J, Lin Z, Gao Y, Yao T. Downregulation of long noncoding RNA MEG3 is associated with poor prognosis and promoter hypermethylation in cervical cancer. J Exp Clin Cancer Res. 2017;36(1):5. doi:10.1186/s13046-016-0472-2
5. Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–6043. doi:10.1158/0008-5472.CAN-06-0561
6. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. doi:10.1038/ncomms11215
7. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
8. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
9. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi:10.1016/j.molcel.2013.08.017
10. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
11. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
12. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
13. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
14. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
15. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi:10.1080/15476286.2016.1220473
16. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi:10.1038/cr.2017.31
17. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
18. Rong X, Gao W, Yang X, Guo J. Downregulation of hsa_circ_0007534 restricts the proliferation and invasion of cervical cancer through regulating miR-498/BMI-1 signaling. Life Sci. 2019;235:116785. doi:10.1016/j.lfs.2019.116785
19. Chen T, Shao S, Li W, Liu Y, Cao Y. The circular RNA hsa-circ-0072309 plays anti-tumour roles by sponging miR-100 through the deactivation of PI3K/AKT and mTOR pathways in the renal carcinoma cell lines. Artif Cells Nanomed Biotechnol. 2019;47(1):3638–3648. doi:10.1080/21691401.2019.1657873
20. Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am J Cancer Res. 2015;5(5):1602–1609.
21. Zeng L, Liao Q, Zou Z, et al. Long non-coding RNA XLOC_006753 promotes the development of multidrug resistance in gastric cancer cells through the PI3K/AKT/mTOR signaling pathway. Cell Physiol Biochem. 2018;51(3):1221–1236. doi:10.1159/000495499
22. Hare SH, Harvey AJ. mTOR function and therapeutic targeting in breast cancer. Am J Cancer Res. 2017;7(3):383–404.
23. Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60(4):855–865. doi:10.1016/j.jhep.2013.11.031
24. Bahrami A, Hasanzadeh M, Hassanian SM, et al. The potential value of the PI3K/Akt/mTOR signaling pathway for assessing prognosis in cervical cancer and as a target for therapy. J Cell Biochem. 2017;118(12):4163–4169. doi:10.1002/jcb.v118.12
25. de Melo AC, Paulino E, Garces AH. A review of mTOR pathway inhibitors in gynecologic cancer. Oxid Med Cell Longev. 2017;2017:4809751. doi:10.1155/2017/4809751
26. Xie J, Wang X, Proud CG. mTOR inhibitors in cancer therapy. F1000Research. 2016;5.
27. Assad DX, Elias ST, Melo AC, Ferreira CG, De Luca Canto G, Guerra EN. Potential impact of mTOR inhibitors on cervical squamous cell carcinoma: a systematic review. Oncol Lett. 2016;12(5):4107–4116. doi:10.3892/ol.2016.5157
28. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027
29. Dai Y, Li D, Chen X, et al. Circular RNA Myosin Light Chain Kinase (MYLK) promotes prostate cancer progression through modulating Mir-29a expression. Med Sci Monit. 2018;24:3462–3471. doi:10.12659/MSM.908009
30. Duan X, Shen N, Chen J, Wang J, Zhu Q, Zhai Z. Circular RNA MYLK serves as an oncogene to promote cancer progression via microRNA-195/cyclin D1 axis in laryngeal squamous cell carcinoma. Biosci Rep. 2019;39(9). doi:10.1042/BSR20190227
31. Caron E, Ghosh S, Matsuoka Y, et al. A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2010;6(1):453. doi:10.1038/msb.2010.108
32. Yang H, Jiang X, Li B, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552(7685):368–373. doi:10.1038/nature25023
33. Hu C, Wang Y, Li A, Zhang J, Xue F, Zhu L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol. 2019;234(6):9225–9232. doi:10.1002/jcp.v234.6
34. Jiao J, Zhang T, Jiao X, et al. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J Cell Physiol. 2020;235(2):1287–1295.
35. Li Z, Hu Y, Zeng Q, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 2019;19(1):211. doi:10.1186/s12935-019-0926-7
36. Armijo ME, Campos T, Fuentes-Villalobos F, Palma ME, Pincheira R, Castro AF. Rheb signaling and tumorigenesis: mTORC1 and new horizons. Int J Cancer. 2016;138(8):1815–1823. doi:10.1002/ijc.v138.8
37. Tian Y, Shen L, Li F, Yang J, Wan X, Ouyang M. Silencing of RHEB inhibits cell proliferation and promotes apoptosis in colorectal cancer cells via inhibition of the mTOR signaling pathway. J Cell Physiol. 2020;235(1):442–453.
38. Lu ZH, Shvartsman MB, Lee AY, et al. Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res. 2010;70(8):3287–3298. doi:10.1158/0008-5472.CAN-09-3467
39. Hanrahan J, Blenis J. Rheb activation of mTOR and S6K1 signaling. Methods Enzymol. 2006;407:542–555.
40. Xie G, Wang Z, Chen Y, et al. Dual blocking of PI3K and mTOR signaling by NVP-BEZ235 inhibits proliferation in cervical carcinoma cells and enhances therapeutic response. Cancer Lett. 2017;388:12–20. doi:10.1016/j.canlet.2016.11.024
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.