Back to Journals » Cancer Management and Research » Volume 13
CircRNA hsa_circ_0005909 Promotes Cell Proliferation of Osteosarcoma Cells by Targeting miR-338-3p/HMGA1 Axis
Authors Zhang C, Na N, Liu L, Qiu Y
Received 3 October 2020
Accepted for publication 22 December 2020
Published 27 January 2021 Volume 2021:13 Pages 795—803
DOI https://doi.org/10.2147/CMAR.S285118
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Beicheng Sun
Cailong Zhang,1 Na Na,2 Li Liu,3 Yingzhu Qiu4
1Department of Arthrology Surgery, The Affiliated Hospital of Qingdao University, Qingdao 266000, People’s Republic of China; 2Department of Obstetrics, Qingdao Eighth People’s Hospital, Qingdao 266000, People’s Republic of China; 3Department of Stereotactic Radiotherapy, Qingdao Central Hospital, Qingdao 266000, People’s Republic of China; 4Department of Spine Surgery, Zibo Central Hospital, Zibo 255000, People’s Republic of China
Correspondence: Yingzhu Qiu
Department of Spine Surgery, Zibo Central Hospital, 56 West Gongqingtuan Road, Zhangdian District, Zibo 255000, People’s Republic of China
Email [email protected]
Objective: Osteosarcoma (OS) is the most common malignant bone tumor in the pediatric population. The main goal of this study is to investigate the role of hsa_circ_0005909 and the underlying signaling pathway involved in OS.
Methods: Cell proliferation was measured using a CCK-8 assay kit and clone formation assay. Change of RNA and protein expression was determined using RNA extract and quantitative real time PCR (RT-qPCR) assay and Western blotting, respectively. CircInteractome was used to predict the target of circRNA and starBase v2.0 was used to predict the target of miRNAs. Luciferase assay was used to confirm the predicted results from CircInteractome, starBase v2.0, and MirTarget2.
Results: Expression of circ_0005909 was upregulated in both OS tissues and cell lines. The predicted results from CircInteractome, starBase v2.0, and MirTarget2 demonstrated that circ_0005909 could sponge miR-338-3p and that HGMA1 was the direct target of miR-338-3p. Cell viability and cell clones were inhibited by knockdown of circ_0005909 but increased by dual inhibition of circ_0005909 and miR-338-3p. Phosphorylation of ERK, Akt, and PI3K was inhibited by sh-circ_0005909, while this inhibition was repressed by co-transfection of sh-circ_0005909 and HGMA1.
Conclusion: Expression of circ_0005909 was upregulated in both OS tissues and cell lines which upregulated expression of HGMA1 through sponging miR-338-3p, resulting in the activation of MAPK-ERK and PI3K-Akt signaling pathways to promote the development of OS.
Keywords: hsa_circ_0005909, osteosarcoma, miR-338-3p, HMGA1
Introduction
Osteosarcoma (OS) is a class of primary malignant bone tumor, which was most common in children and adolescents.1 The 5-year survival was approximately 65–70% in OS patients with localized disease while it was only 19–30% for metastatic disease.2 Currently, the standard systematic chemotherapeutic drugs for OS are: high-dose methotrexate with leucovorin rescue (HDMTX), doxorubicin, and cisplatin (MAP).3,4 With the complete surgical resection, this adjuvant chemotherapy achieved 60–70% of 5-year event-free survival in patients with non-metastatic disease.3 However, these three drugs are all cytotoxic agents with many side-effects,5 including cardiotoxicity.6–8 Therefore, new treatment regimens should be explored to improve the outcomes and life quality of OS patients.
Many signaling pathways have been involved in the development of OS.9 For example, the PI3K/Akt signaling pathway was excessively activated in OS and promoted tumorigenesis, cell cycle progression, and inhibition of cell apoptosis, leading to the development of OS.10 The MAPK/ERK pathway is also an important signaling pathway, over-activation of the MAPK/ERK pathway was enhance cell proliferation and accelerate drug resistance in OS.9 Circular RNAs (circRNAs) are a type of long, non-coding RNA which is resistant to endonuclease due to its closed loop structure.11,12 circRNAs contain a conserved miRNA binding site, which resulted in repression of target miRNA activity.13 Unlike circRNAs, microRNAs (miRNAs) are a class of small, linear RNAs with only 18–25 nucleotides (nt) in length.14 miRNAs are also non-coding RNAs which repress the translational process via binding to the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs.15 Acting as a competing endogenous RNA (ceRNA), circRNAs can protect the target mRNA from degradation to prevent translational repression.16 Therefore, new technologies for target therapy and gene therapy should be considered for cancer therapy.17
Recent studies have reported that both circRNAs and miRNAs are involved in the regulation of tumor growth and metastasis.18 As reported by Li et al,19 upregulation of circ-0001785 could sponge miR-1200 to induce the overexpression of HOXB2, leading to the progression of OS. Acting as a ceRNA, circ_0102049 could upregulate MDM2 expression through targeting miR-1304-5p, promoting cell growth in OS.20 On the contrary, circ_HIPK3 was downregulated in OS and overexpression of circ_HIPK3 prevented cell proliferation, migration, and invasion in OS, demonstrating the inhibitory effects on the pathogenesis of OS.21 Circ_0005909 promoted the progression of OS through upregulating HMGB1 via sponging miR-936.22 These data suggest that circRNAs play important roles in cancer diagnosis and therapy through sponging target miRNAs. The main goal of this study is to investigate the role of hsa_circ_0005909 and the underlying signaling pathway involved in OS.
Methods
Human Osteosarcoma Tissue Samples
Human OS tissue samples and their adjacent non-cancerous bone tissues (n=15) were collected during surgery from November 25, 2019 to January 15, 2020 at Zibo Central Hospital and written informed consent was obtained from all patients (Table 1). All the OS tissues and non-cancerous bone tissues were confirmed by pathologists. The collected sample was frozen by liquid nitrogen and stored at −80°C. The protocol of this research project has been approved by the Ethics Committee of Zibo Central Hospital (Approval Number: K Y-E-2019-11-20) and was in accordance with the Declaration of Helsinki.
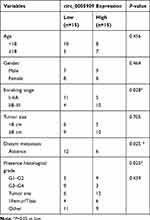 |
Table 1 Patient Characteristics |
Cell Culture
Human OS cell lines (MG63, HOS, 143B, and U2OS cells) and human osteoprogenitor cell line hFOB1.19 cells were obtained from American Type Culture Collection (ATCC, USA). And 143B cells have been authenticated using STR methods. All cells were incubated in Dulbecco’s modified Eagles’ medium (DMEM, HyClone, USA) supplemented with 1% penicillin/streptomycin (Invitrogen, USA) and 10% fetal bovine serum (FBS, Invitrogen, USA) under 37°C with 95% air and 5% CO2 in a humidified incubator.
For transfection experiments, cells were cultured in sterile 12-well plates at the density of 1×106 cells/well. miR-338-3p mimics, miR-338-3p inhibitors, small hairpin RNA of circ_0005909 (sh-circ_0005909), and their negative controls were purchased from Guangzhou Bersin Biotechnology Co. Ltd (BersinBio, China) and transfected into the cultured cells with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturers’ protocols. The transfected cells were cultured under 37°C with 95% air and 5% CO2 for 24 hours for further experiments and analysis.
RNA Extract and Quantitative Real Time PCR (RT-qPCR) Assay
After treatment, total RNA was extracted with Trizol reagent (TaKaRa, China) according to the manufacturer’s instructions. Total RNA (500 ng) was reverse transcribed to cDNA with the PrimeScript RT Master Mix (TaKaRa, China). The relative RNA expression was examined using SYBR® Green Quantitative RT-qPCR Kit (Sigma-Aldrich, USA) and calculated by means of the 2−ΔΔCt method. The primer sequences (Sigma-Aldrich, USA) are shown in Table 2.
 |
Table 2 Primer Sequences for RT-qPCR |
RNase R Treatment and Colony Formation
In total, 5 μg of total RNA was incubated with or without 3 U/μg of RNase R (Epicentre Biotechnologies, USA) at 37°C for 15 minutes. Expression of RNA was subsequently purified and extracted by phenol-chloroform and then subjected to qRT‐PCR.
CCK-8 Assay and Clone Formation Assay
After treatment, cell proliferation was examined at 0, 24, 48, and 72 hours by Cell Counting Kit-8 (CCK-8) assay (Dojido, Japan) in a 96-well plate according to the manufacturer’s instructions. Briefly, 10 µL of CCK-8 solution was added to each well. The solution was then measured spectrophotometrically at 450 nm after 2 hours incubation at 37°C and the optical density (OD) value was recorded.
For clone formation assay, MG63 and 143B cells were mixed with 0.4% of agarose and seeded into 12-well plates. Then 2 mL of culture medium was added to each well and incubated at 37°C with 5% CO2 for 7 days. MG63 and 143B cells were fixed with 10% methanol and stained with 0.1% crystal violet. The cell colonies were counted and photographed under a microscope (Olympus, Japan).
Construct of Plasmid and Luciferase Assay
This protocol followed a published paper.23 Briefly, the full-length of HMGA1 3ʹ-UTR or circ_0005909 containing wild type (wt) and scrambled (mut) miR-338-3p binding sequence were inserted downstream of the firefly luciferase gene in psiCHECK2 to generate the psiCHECK2-HMGA1 3ʹUTR-wt or circ_0005909-mut plasmid and psiCHECK2-HMGA1 3ʹUTR-mut plasmid or circ_0005909-mut, respectively. The wt and mut plasmids were co-transfected into H293T cells with negative control, miR-338-3p mimics or their negative control (miR-NC) with control Renilla luciferase expression plasmid using Lipofectamine 2000. After 24 hours, relative luciferase activity was measure using the Dual-luciferase reporter Assay System (Promega, USA) according to the manufacturer’s instructions.
Western Blotting
The treated cells were suspended and lysed with RIPA lysis buffer (Beyotime, China) to extract the protein. The cytoplasm and nuclear proteins were extracted using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher, USA) according to the manufacturer’s protocol. Protein samples were loaded into sodium dodecyl sulfate polyacrylamide gel (7.5%) and separated by electrophoresis. The proteins were then transferred to PVDF membranes. The membranes were blocked by 5% fat-free milk for 1 hour at room temperature and probed with proper primary antibodies overnight at 4 °C followed by incubating with secondary antibodies for 2 hours at room temperature. The protein signals were detected using Pierce™ ECL Western Blotting Substrate (Thermo Fisher, USA).
Bioinformatics Analysis
CircInteractome (https://circinteractome.nia.nih.gov/) was used to predict miR-338-3p binding site in the circ_0005909. MirTarget2 (http://mirdb.org/) and starBase v2.0 (http://starbase.sysu.edu.cn/) was used to predict the potential miR-338-3p binding sites to corresponding gene HMGA1 3ʹUTR to study the possible crossing network among circRNA, miRNA, and target gene.
Statistical Analysis
GraphPad Prism 8.1.0 (GraphPad, USA) was used to perform the statistical analysis in this study. Comparisons between two groups were analyzed using Student’s t-test. Comparisons among more than two groups were performed using one-factor analysis of variance followed by the Bonferroni’s post hoc tests. The correlation between circ_0005909 and miR-338-3p was evaluated using Spearman’s rank correlation test. All data was presented as mean±standard error of means (SEM). P<0.05 represents statistical significance.
Results
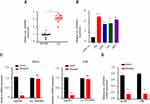
hsa_circ_0005909 Was Upregulated in OS
Expression of circ_0005909 was upregulated in OS tissues compared with their adjacent normal tissues (Figure 1A). This upregulation of circ_0005909 was also observed in human OS cell lines, MG63, HOS, 143B, and U2OS cells, compared with human osteoprogenitor cell line hFOB1.19 cells (Figure 1B). RNase R treatment was used to confirm the circular characteristics of circ_0005909. The results demonstrated that circ_0005909 did not show any significant change in the RNase R treatment group while the expression of the linear control RNA, GAPDH, was significantly reduced after RNase R treatment compared with the mock group in both MG63 and 143B cells (Figure 1C). The expression of circ_0005909 was higher in plasma than that in the nucleus in both MG63 and 143B cells (Figure 1D).
circ_0005909 Acted as a Molecular Sponge of miR-338-3p in OS Cells
CircInteractome was used to predict the target miRNA of circ_0005909. The results showed that there was a complementary sequence between circ_0005909 and miR-338-3p (Figure 2A). Luciferase activity was reduced in cells co-transfected with circ_0005909-wt plasmid and miR-338-3p mimics compared with a negative control group and circ_0005909-mut group (Figure 2B). The expression of miR-338 was decreased in OS tissues compared with their adjacent normal tissue, which is negatively correlated with the expression of circ_0005909 (Figure 2C). RNA pull-down assay showed that higher expression of miR-338-3p enriched more circ_0005909 compared with the negative control group in both MG63 and 143B cells (Figure 2D).
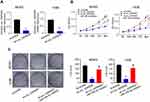
Knockdown of circ_0005909 Repressed Proliferation in OS Cells via Regulating miR-338-3p
Sh-circ_0005909 was used to downregulate the expression of circ_0005909 in MG63 and 143B cells. The expression of circ_0005909 was significantly reduced in cells transfected with sh-circ_0005909 compared with cells transfected with scramble RNAs in both MG63 and 143B cells (Figure 3A). Results from CCK-8 assay demonstrated that cell viability was significantly reduced by inhibition of circ_0005909 while this reduction of cell viability was reversed by dual inhibition of circ_0005909 and miR-338-3p (Figure 3B). The number of cell clone spots was significantly decreased by sh-circ_0005909 while this decrease of cell clones was attenuated by dual inhibition of circ_0005909 and miR-338-3p (Figure 3C).
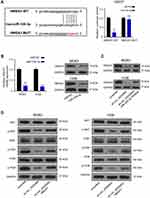
HMGA1 Was the Direct Target of miR-338-3p
The prediction results from starBase v2.0 and MirTarget2 indicated that miR-338-3p could bind to the 3ʹ-UTR of HMGA1 (Figure 4A). Luciferase activity was reduced in cells co-transfected with HMGA1-wt plasmid and miR-338-3p mimics compared with the negative control group and HMGA1-mut group (Figure 4A). Both mRNA and protein expression of HMGA1 was repressed in MG63 and 143B cells transfected with miR-338-3p mimics compared with those transfected with negative control (Figure 4B). The protein expression of HMGA1 was downregulated by sh-circ_0005909 while this downregulation was prevented by dual inhibition of circ_0005909 and miR-338-3p in both MG63 and 143B cells (Figure 4C). The phosphorylation of ERK, Akt, and PI3K was downregulated by sh-circ_0005909, while this inhibition was repressed by co-transfection of sh-circ_0005909 and HGMA1 in both MG63 and 143B cells (Figure 4D).
Discussion
OS is an aggressive bone tumor in the pediatric population with a poor prognostic outcome in patients with metastatic disease.2 The pathogenesis and new mechanisms underwent deep investigation to improve the prognosis and prolongthe patients’ life. In this study, circ_0005909 was overexpressed in both tissues and cell lines of OS. Further experiments have found that circ_0005909 upregulated expression of HGMA1 through sponging miR-338-3p, which activated MAPK-ERK and PI3K-Akt signaling pathways and promoted the development of OS, providing a new understanding and therapeutic target for treatment of OS in the future.
Many circRNAs have involved into the pathogenesis of OS and have become an important diagnostic and prognostic indicator in OS.5,24 In the present study, overexpression of circ_0005909 was observed in both OS tissues and cell lines, which is consistent with previous studies.22,25 Among the tested OS cell lines, MG63 and 143B cells demonstrated higher expression of circ_0005909 than in other OS cell lines. Thus, MG63 and 143B cells were chosen for further study. The data of this study have shown that circ_0005909 was overexpressed in OS. Knockdown of circ_0005909 reduced cell proliferation in OS cell lines, indicating that circ_0005909 contributed to the development and progression of OS. A previous study has revealed that circ_0005909 could upregulate HMGB1 via targeting miR-936 in OS.22 However, results from this study manifested that circ_0005909 could also sponge miR-338-3p to increase cell proliferation in OS. This could be explained by “off-target” effects of miRNAs, which means each miRNA could bind to hundreds of target mRNAs or circRNAs with partial sequence match to regulate multiple biological processes.26,27
Results from the starBase v2.0 and MirTarget2, and luciferase assay have confirmed that HMGA1 was the direct target of miR-338-3p. HMGA1 is a family member of the high mobility group A (HMGA) proteins.28 HMGA1 is upregulated in highly proliferative cells, and is closely associated with aggressiveness and poor prognosis of cancer.28 In this study, expression of miR-338-3p was negatively associated with expression of circ_0005909 in OS, and overexpression of miR-338-3p downregulated HMGA1. Thus, there would be a positive correlation between the expression of HMGA1 and circ_005909. This evidence proved that circ_005909 upregulated expression of HMGA1 via targeting miR-338-3p, leading to the development and progression of OS.
Both MAPK-ERK1/2 signaling pathway and PI3K-Akt signaling pathway were overactivated in OS cells compared with normal cells.9 And many studies have revealed that activation of these two pathways contributed to cell proliferation, which resulted in tumor growth and metastasis.10,29 HMGA1 has been reported to activate the MAPK-ERK1/2 signaling pathway and PI3K-Akt signaling pathway.30,31 In the present study, phosphorylation of ERK, PI3K, and Akt was inhibited by knockdown of circ_0005909, suggesting that circ_0005909 involved into the activation of MAPK-ERK and PI3K-Akt signaling pathways. However, the downregulation of phosphorylated ERK, PI3K, and Akt was reversed by overexpression of HMGA1. Taken together, these results demonstrated that circ_0005909 could promote the activation of MAPK-ERK and PI3K-Akt signaling pathways, which was mediated by upregulation of HMGA1, thus contributing to the pathogenesis of OS. Some limitations existed in this study: 1) The data did not compare the phosphorylation of ERK, Akt, and PI3K in normal cells and in OS cell lines, but it is well known that the MAPK-ERK1/2 signaling pathway and PI3K-Akt signaling pathway was overactivated in OS cells compared with normal cells;9 2) The results should be further validated using an OS animal model in future studies; 3) The sample size was very low if this is a clinical study. However, this was a basic scientific study to figure out the effects of circ_0005909 in osteosarcoma and the underlying mechanism. Therefore, the sample size is enough to demonstrate the change of circ_0005909 expression in OS in this basic scientific study.
In summary, the data of the present study have shown that the expression of circ_0005909 was upregulated in both OS tissues and cell lines. Acting as ceRNA, circ_0005909 upregulated expression of HGMA1 through sponging miR-338-3p, which enhanced the activation of the MAPK-ERK and PI3K-Akt signaling pathways to promote the development of OS. These results suggested that circ_0005909 and miR-338-3p can be effective biomarkers for the diagnosis and prognosis of OS and that further investigation of new drugs for OS could focus on targeting HGMA1 and MAPK-ERK and PI3K-Akt signaling pathways.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflict of interests.
References
1. Kumar R, Kumar M, Malhotra K, Patel S. Primary osteosarcoma in the elderly revisited: current concepts in diagnosis and treatment. Curr Oncol Rep. 2018;20(2):13. doi:10.1007/s11912-018-0658-1
2. Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28(1):26–33. doi:10.1097/MOP.0000000000000298
3. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. doi:10.1080/14737140.2018.1413939
4. Mir SM, Yousefi B, Marjani A, Rahimi M, Qujeq D. The sensitization of melatonin in osteosarcoma cells by suppression of anti-apoptotic proteins. Pharm Sci. 2020;26(2):159–164. doi:10.34172/PS.2020.3
5. Soghli N, Qujeq D, Yousefi T, Soghli N. The regulatory functions of circular RNAs in osteosarcoma. Genomics. 2020;112(4):2845–2856. doi:10.1016/j.ygeno.2020.03.024
6. Gorini S, De Angelis A, Berrino L, Malara N, Rosano G, Ferraro E. Chemotherapeutic drugs and mitochondrial dysfunction: focus on doxorubicin, trastuzumab, and sunitinib. Oxid Med Cell Longev. 2018;2018:7582730. doi:10.1155/2018/7582730
7. Shea B, Swinden MV, Tanjong Ghogomu E, et al. Folic acid and folinic acid for reducing side effects in patients receiving methotrexate for rheumatoid arthritis. Cochrane Database Syst Rev. 2013;2013(5):Cd000951.
8. Barabas K, Milner R, Lurie D, Adin C. Cisplatin: a review of toxicities and therapeutic applications. Vet Comp Oncol. 2008;6(1):1–18. doi:10.1111/j.1476-5829.2007.00142.x
9. Czarnecka AM, Synoradzki K, Firlej W, et al. Molecular biology of osteosarcoma. Cancers (Basel). 2020;12(8):2130. doi:10.3390/cancers12082130
10. Zhang J, Yu XH, Yan YG, Wang C, Wang WJ. PI3K/Akt signaling in osteosarcoma. Clin Chim Acta. 2015;444:182–192. doi:10.1016/j.cca.2014.12.041
11. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
12. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559
13. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
14. Rajman M, Schratt G. MicroRNAs in neural development: from master regulators to fine-tuners. Development. 2017;144(13):2310–2322. doi:10.1242/dev.144337
15. Zhang Y, Sun X, Icli B, Feinberg MW. Emerging roles for MicroRNAs in diabetic microvascular disease: novel targets for therapy. Endocr Rev. 2017;38(2):145–168.
16. Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875–2889. doi:10.1007/s00432-019-03045-4
17. Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5(1):019–0089.
18. Skrzypek K, Majka M. Interplay among SNAIL transcription factor, MicroRNAs, long non-coding RNAs, and circular RNAs in the regulation of tumor growth and metastasis. Cancers. 2020;12(1):209. doi:10.3390/cancers12010209
19. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle. 2019;18(11):1281–1291. doi:10.1080/15384101.2019.1618127
20. Jin Y, Li L, Zhu T, Liu G. Circular RNA circ_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR-1304-5p in osteosarcoma. Pathol Res Pract. 2019;215(12):152688. doi:10.1016/j.prp.2019.152688
21. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi:10.7150/jca.24619
22. Ding S, Zhang G, Gao Y, Chen S, Cao C. Circular RNA hsa_circ_0005909 modulates osteosarcoma progression via the miR-936/HMGB1 axis. Cancer Cell Int. 2020;20:305. doi:10.1186/s12935-020-01399-1
23. Xu X-W, Zheng B-A, Hu Z-M, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683. doi:10.18632/oncotarget.21748
24. Zhang Y, Li J, Wang Y, Jing J, Li J. The roles of circular RNAs in osteosarcoma. Med Sci Monit. 2019;25:6378–6382. doi:10.12659/MSM.915559
25. Chen G, Wang Q, Yang Q, et al. Circular RNAs hsa_circ_0032462, hsa_circ_0028173, hsa_circ_0005909 are predicted to promote CADM1 expression by functioning as miRNAs sponge in human osteosarcoma. PLoS One. 2018;13(8):e0202896. doi:10.1371/journal.pone.0202896
26. Seok H, Lee H, Jang ES, Chi SW. Evaluation and control of miRNA-like off-target repression for RNA interference. Cell Mol Life Sci. 2018;75(5):797–814. doi:10.1007/s00018-017-2656-0
27. Lam JKW, Chow MYT, Zhang Y, Leung SWS. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4(9):e252. doi:10.1038/mtna.2015.23
28. Sumter TF, Xian L, Huso T, et al. The high mobility group A1 (HMGA1) transcriptome in cancer and development. Curr Mol Med. 2016;16(4):353–393. doi:10.2174/1566524016666160316152147
29. Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. doi:10.1517/14728222.2011.645805
30. Treff NR, Pouchnik D, Dement GA, Britt RL, Reeves R. High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene. 2004;23(3):777–785. doi:10.1038/sj.onc.1207167
31. Liau -S-S, Jazag A, Whang EE. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66(24):11613. doi:10.1158/0008-5472.CAN-06-1460
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.