Back to Journals » Cancer Management and Research » Volume 13
CircRNA circ-OGDH (hsa_circ_0003340) Acts as a ceRNA to Regulate Glutamine Metabolism and Esophageal Squamous Cell Carcinoma Progression by the miR-615-5p/PDX1 Axis
Authors Liang Z, Zhao B, Hou J, Zheng J, Xin G
Received 2 November 2020
Accepted for publication 1 March 2021
Published 7 April 2021 Volume 2021:13 Pages 3041—3053
DOI https://doi.org/10.2147/CMAR.S290088
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Zongying Liang, Baoshan Zhao, Jishen Hou, Jingxiong Zheng, Guohua Xin
Department of Thoracic Surgery, The Affiliated Hospital of Chengde Medical University, Chengde, 067000, People’s Republic of China
Correspondence: Guohua Xin
Department of Thoracic Surgery, The Affiliated Hospital of Chengde Medical University, No. 36, Nanyingzi Street, Shuangqiao District, Chengde, 067000, People’s Republic of China
Email [email protected]
Background: Circular RNA hsa_circ_0003340 (circ-OGDH) has been uncovered to be involved in esophageal squamous cell carcinoma (ESCC) progression. However, the mechanism by which circ-OGDH regulates ESCC progression is unclear.
Methods: Expression levels of circ-OGDH, microRNA (miR)-615-5p, and PDX1 (pancreatic and duodenal homeobox 1) mRNA were evaluated with quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation, apoptosis, migration, invasion, and cell cycle progression of ESCC cells were analyzed by MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide), colony formation, flow cytometry, and transwell assays. Measurement of glutamine consumption, α-KG (α-ketoglutarate) production, and ATP (Adenosine Triphosphate) content using corresponding kits. Protein levels were analyzed by Western blotting. The targeting relationship between circ-OGDH or PDX1 and miR-615-5p was verified by dual-luciferase reporter and RNA immunoprecipitation (RIP) assays. The function of circ-OGDH in ESCC was confirmed by animal experiments.
Results: Circ-OGDH was upregulated in ESCC. Circ-OGDH inhibition reduced ESCC growth in vivo and accelerated cell apoptosis, cell cycle arrest, repressed cell proliferation, migration, invasion, and reduced cell glutamine metabolism in ESCC cells in vitro. MiR-615-5p was downregulated in ESCC, while PDX1 had an opposite result. Circ-OGDH sponged miR-615-5p to regulate PDX1 expression. MiR-615-5p inhibitor neutralized the repressive effect of circ-OGDH knockdown on malignancy and glutamine metabolism of ESCC cells. PDX1 overexpression counteracted the inhibitory impact of miR-615-5p mimic on malignancy and glutamine metabolism of ESCC cells.
Conclusion: Circ-OGDH sponged miR-615-5p to elevate PDX1 expression, thus elevating glutamine metabolism and promoting tumor growth in ESCC. The study offered evidence to support circ-OGDH as a promising target for ESCC therapy.
Keywords: ESCC, circ-OGDH, miR-615-5p, PDX1, glutamine
Introduction
Esophageal cancer ranks sixth among the causes of cancer deaths worldwide.1 Esophageal squamous cell carcinoma (ESCC), a major subtype of esophageal cancer, accounts for approximately 90% of all esophageal cancer cases in developing countries, including China.2 At present, esophagectomy combined with radical mediastinal lymph node dissection is still the standard treatment for ESCC.3 However, the probability of ESCC recurrence is still high after radical surgery.4,5 Therefore, it is essential to develop new ESCC treatment strategies to improve the prognosis of ESCC.
Unlike normal cells, cancer cells reprogram their metabolic pathways to meet their energy requirements.6 Glutamine, the most abundant nutrient in plasma, is responsible for most of the carbon and nitrogen metabolism in cells. Glutaminase can convert glutamine into α-ketoglutarate (α-KG), which participates in the tricarboxylic acid (TCA) cycle to provide energy for cells.7 Many studies have proved that glutamine metabolism is involved in macromolecule biosynthesis, regulating signaling pathways, and maintaining redox homeostasis in cancer cell, thereby facilitated cancer cell survival and proliferation.8 Also, targeting glutamine metabolism offers a promising direction for the development of cancer treatment strategies.9 Accordingly, it is very vital to survey the mechanism of glutamine metabolism related to tumor progression.
Circular RNAs (circRNAs) are a type of endogenous biomolecules with closed loop structures.10 They have cell-specific and tissue-specific expression patterns and resist to exonuclease RNase R digestion.11 Recent studies have shown that circRNAs can function as transcription regulators, protein binding partners, and microRNA (miR) sponges.12 Moreover, accumulated evidence has proved that circRNAs participate in the progression of many human diseases, including cancer. For example, circRNA circ-DONSON facilitated gastric cancer growth by initiating SOX4 expression via recruiting the NURF complex.13 CircRNA circ-DOXP1 sponged miR-370 and increased PKLR expression, resulting in accelerating Warburg effect and tumor growth in gallbladder cancer.14 CircRNA circ-OGDH (circ-OGDH) (circBase ID: hsa_circ_0003340), located at chr7: 44684925-44687358, is a circRNA with a length of 295 bp produced by reverse splicing of the OGDH gene (http://www.circbank.cn/search.html?selectValue=hsa_circOGDH_007). Report of Hou et al indicated that circ-OGDH acted as an oncogene in ESCC.15 Nevertheless, whether circ-OGDH regulates glutamine metabolism in ESCC progression is unclear.
Hence, this study aimed to explore the mechanism of circ-OGDH regulating glutamine metabolism in ESCC progression. Our results proved that circ-OGDH elevated glutamine metabolism and promoted ESCC growth by regulation of the miR-615-5p/PDX1 (pancreatic and duodenal homeobox 1) axis.
Materials and Methods
Case Selection
The protocols of the study were approved by the Ethics Committee of the Affiliated Hospital of Chengde Medical University and conducted in line with the Declaration of Helsinki. 34 pairs of ESCC tissues and para-carcinoma tissues were collected from patients who underwent ESCC surgical resection at the Affiliated Hospital of Chengde Medical University. All enrolled patients signed written informed consents, and none of them had received chemotherapy or radiotherapy prior to surgery.
Cell Culture
Human normal esophageal epithelial cell line (HEEC) and ESCC cell lines (TE-1 and Eca-109) were obtained from Bena Culture Collection (Suzhou, China). HEEC and Eca-109 were maintained in the DMEM (Dulbecco’s modified Eagle’s medium) (Sigma, St Louis, MO, USA) in a humidified chamber at 37°C with 5% CO2, whereas the TE-1 cells were maintained in RPMI (Roswell Park Memorial Institute)-1640 medium (Sigma). These mediums were supplemented with 10% FBS (Fetal Bovine Serum) (Sigma) and 1% streptomycin/penicillin (Sigma).
Cell Transfection
The RNA oligonucleotides used in this study were purchased from GenePharma (Shanghai, China), including small interference (si) RNA against circ-OGDH (si-circ-OGDH#1 and si-circ-OGDH#2) and its matched negative control (si-NC), miR-615-5p mimic (miR-615-5p), negative control matched with miR mimic (miR-NC), miR-615-5p inhibitor (anti-miR-615-5p), and negative control corresponding miR inhibitor (anti-miR-NC). Overexpression plasmids of pCD5-ciR-circ-OGDH (circ-OGDH) and pcDNA-PDX1 (PDX1) were constructed using empty pCD5-ciR (Vector) (Geneseed, Guangzhou, China) and pcDNA (Thermo Fisher Scientific) vectors. Transient transfection was carried out using the Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA) or Lipofectamine 3000 reagent (Thermo Fisher Scientific).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA isolation was conducted with the RNAiso plus (Takara, Dalian, China). The concentration and integrity of total RNA were evaluated using the SmartSpec Plus spectrophotometer (Bio-Rad, Hercules, CA, USA) and agarose gel electrophoresis (1%). To analyze the stability of circ-OGDH, total RNA derived from ESCC cells was digested with RNase R (3U/ug, Epicentre Technologies, Madison, WI, USA) at 37°C for 20 min, and DEPC (diethyl pyrocarbonate)-treated water (Thermo Fisher Scientific) was utilized as a negative control. QRT-PCR was carried out with the Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) on the Light Cycler 480 System (Roche, Basel, Switzerland). The complementary DNA (cDNA) was produced using the miRcut Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China) or ReverTra Ace™ qPCR RT Kit (Toyobo, Osaka, Japan). The primer sequences were displayed in Table 1. Relative expression was calculated by the 2−ΔΔCt method and normalized to U6 or β-actin.
 |
Table 1 Primer Sequences Used for qRT-PCR |
Cell Viability Analysis
The 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay was conducted using the Cell Proliferation Kit I (Roche) to assess cell viability. In short, ESCC cells were transfected with oligonucleotides and/or plasmids and then incubated onto 96-well plates for 48 h. Thereafter, the MTT reagent (5 mg/mL, 10 μL) was added to each well and incubated for 4 h. Subsequently, the solubilization solution (100 μL) was supplemented to dissolve the purple crystals. The OD (optical density) value of the solution at 570 nm was measured with a BioTek™ 800TS Absorbance Reader (BioTek, Winooski, VT, USA).
Colony Formation Assay
After transfection for 24 h, ESCC cells (1 × 103) were seeded into 6-well plates containing complete medium (2 mL) and incubated for 10 days. Then, the cells were washed with PBS (phosphate buffer saline) (Sigma) and fixed with 4% paraformaldehyde (Santa Cruz Biotechnology, Santa Cruz, CA, USA). After dying with 0.1% crystal violet (Santa Cruz Biotechnology), the number of colonies (> 50 cells) was counted using a Nikon microscope (Nikon Eclipse E600, Nikon Instruments, Melville, NY, USA).
Cell Apoptosis Analysis
Flow cytometry assay was conducted with the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Becton Dickinson, San Jose, CA, USA). After digestion with trypsin (Sigma), the cells (1 × 106) were re-suspended in 1× binding buffer. Then, the cells (1 × 105) were stained with Annexin V-FITC (5 μL) and PI (5 μL). The cells were analyzed using a FACS (fluorescence-activated cell sorting) Calibur Flow Cytometer (Becton Dickinson) with a CellQuest software program (Becton Dickinson).
Cell Migration and Invasion Analysis
Transwell assay was performed using transwell migration chambers (#3422, Costar, Cambridge, MA, USA) or invasion chambers (#354480, Costar). Briefly, ESCC cells re-suspended in cell medium (200 μL) were inoculated into apical chambers after transfection for 24 h. Also, 600 μL medium containing 10% FBS (Thermo Fisher Scientific) was added to basolateral chambers. After culture for a while (24 h), the migrating and invasive cells were fixed with 4% paraformaldehyde (Santa Cruz Biotechnology) and stained with 0.1% crystal violet (Santa Cruz Biotechnology). The cells in 5 random fields were figured using a Nikon microscope (× 100, Nikon Eclipse E600, Nikon Instruments).
Cell Cycle Progression Analysis
The cell cycle progression was evaluated using the DNA Content Detection Kit (Solarbio, Beijing, China). In short, the cells were digested and washed with trypsin (Sigma) and PBS (Sigma), respectively. Thereafter, the cells (1 × 106) were fixed with 70% pre-cooled ethanol (500 μL), followed by disposing with RNase A (100 μL) at 37°C for 30 min. Subsequently, the cells were stained with PI (400 μL) at 4°C for 30 min. Finally, the cell distribution was evaluated using a FACS (fluorescence-activated cell sorting) Calibur Flow Cytometer (Becton Dickinson) with a CellQuest software program (Becton Dickinson).
Glutamine Consumption, α-KG Production, and ATP (Adenosine Triphosphate) Content Measurements
The level of glutamine in the culture medium was measured with a Glutamine Assay Kit (Bioassay Systems, Hayward, CA, USA). Briefly, the cell medium was mixed with the enzyme after deproteinization with an ultracentrifugal filter. The absorbance at 565 nm was measured with a BioTek™ 800TS Absorbance Reader (BioTek) after incubation for 40 min in the dark. Glutamine consumption was calculated according to the following equation: Glutamine consumption = glutamine in fresh medium-glutamine in cell medium.
The levels of α-KG and ATP in cultured cells were assessed with α-KG colorimetric assay kit (BioVision, Milpitas, CA, USA) or CellTiter-Glo Luminescent Cell Viability Assay kit (Promega, Madison, WI, USA) in accordance with manufacturer’s instructions.
Western Blotting
Total protein was extracted with the RIPA (Radio-Immunoprecipitation Assay) buffer (Thermo Fisher Scientific) and quantified with the QuantiPro™ BCA Assay Kit (Sigma). Total protein was loaded into 10% sodium dodecyl sulfate-polyacrylamide gel (Sigma) and separated by electrophoresis. The separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and then sealed with 5% non-fat milk. Thereafter, the membranes were washed with TBST (Tris Buffered Saline Tween) (Thermo Fisher Scientific) and then incubated with primary antibodies against GLS1 (glutaminase-1) (ab156876, 1:1000, Abcam, Cambridge, MA, USA), PDX1 (ab134150 1:1000, Abcam), and β-actin (ab115777, 1:200, Abcam). Then, the membranes were incubated with an HRP (horseradish peroxidase)-conjugated goat anti-rabbit IgG (ab205718, 1:2000, Abcam) secondary antibody. β-actin was deemed as a loading control. The blots were detected by enhanced chemiluminescence (ECL) substrate (PerkinElmer, Waltham, MA, USA).
Dual-Luciferase Reporter Assay
The binding sites between miR-615-5p and circ-OGDH or PDX1 were predicted using circular RNA interactome and TargetScan. The fragments of wild type (wt) circ-OGDH and 3ʹ untranslated regions (UTR) of PDX1 and their mutant (mut) sequences were inserted into the downstream of the pMIR-REPORT reporter (Applied Biosystems), respectively. ESCC cells were co-transfected with a luciferase reporter containing wt-circ-OGDH, mut-circ-OGDH, wt-PDX1 3ʹUTR, or mut-PDX1 3ʹUTR and miR-NC or miR-615-5p mimic. Thereafter, the cells were lysed and the luciferase activities were assessed with a luciferase assay kit (Biovision) in a TD20/20 Luminometer (Turner Biosystems, Sunnyvale, CA, USA).
RNA Immunoprecipitation (RIP) Assay
RIP analysis was carried out with the Magna RIP kit (Millipore, Bedford, MA, USA) to analyze the specific binding between miR-615-5p and circ-OGDH or PDX1 based on the manufacturer’s instructions. In brief, the lysates of ESCC cells were incubated with immunoprecipitation buffer containing magnetic beads conjugated to Ago2 (argonaute 2) antibody (#10686-1-AP, Thermo Fisher Scientific) or IgG (immunoglobulin G) antibody (#PP64B, Millipore) at 4°C for 6 h, and IgG was used as a control. The enrichment of circ-OGDH or PDX1 and miR-615-5p in purified RNA complexes was examined by qRT-PCR.
Animal Experiments
The animal experiments were ratified by the Animal Ethics Committee of The Affiliated Hospital of Chengde Medical University. 10 female BALB/c nude mice (4–6 weeks old) (Vital River Laboratory, Beijing, China) were randomly divided into 2 groups and injected with TE-1 cells (1×107), which were stably transduced with short hairpin (sh) RNA against circ-OGDH (sh-circ-OGDH) or sh-NC. All mice were housed in pathogen-free conditions and tumor volume was measured once a week. At the end point (5 weeks later), all mice were killed and their tumor tissues were excised, weighed, and conserved at −80°C for subsequent analysis. Animal experiments were conducted in line with the Laboratory animal-Guideline for ethical review of animal welfare of China (permit number: GB/T 35892–2018). The tumor volume was calculated based on the following equation: Volume = (length ×width2)/2.
Statistical Analysis
All experiments were repeated 3 times and each experiment was performed in triplicate. Statistical analysis was executed using GraphPad Prism 7.0 (Graph-Pad Software, La Jolla, CA, USA). All data were expressed as mean ± standard deviation. The normal distribution of the results was confirmed by the Kolmogorov–Smirnov test. The differences between 2 groups or among multiple groups were evaluated with Student’s t-test or one-way ANOVA (analysis of variance) with Turkey’s post hoc test. P < 0.05 was the threshold of significance.
Results
Circ-OGDH Was Overtly Upregulated in ESCC
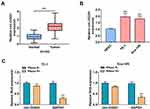
Previous research has revealed that circ-OGDH acts as an oncogene in ESCC.15 To further verify this result, we detected circ-OGDH expression in 34 paired ESCC tissues and para-carcinoma tissues. QRT-PCR showed that circ-OGDH expression was prominently upregulated in ESCC tissues in contrast to para-carcinoma tissues (Figure 1A). As expected, circ-OGDH was highly expressed in ESCC cells (TE-1 and Eca-109) compared with the HEEC cells (Figure 1B). To evaluate the stability of circ-OGDH, we processed RNA from TE-1 and Eca-109 cells with RNase R and observed that circ-OGDH was resistant to RNase R digestion, whereas linear GAPDH was markedly digested after RNase R treatment (Figure 1C). Our data exhibited that circ-OGDH expression was elevated in ESCC.
Circ-OGDH Elevated Glutamine Metabolism and Accelerated Proliferation, Metastasis, and Invasion of ESCC Cells
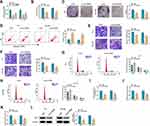
To survey circ-OGDH function in ESCC, we carried out loss-of-function experiments. 2 siRNAs (si-circ-OGDH#1 and si-circ-OGDH#2) against circ-OGDH were used to silence circ-OGDH expression. As exhibited in Figure 2A, circ-OGDH expression was prominently decreased in TE-1 and Eca-109 cells after transfection with si-circ-OGDH#1 or si-circ-OGDH#2, and the si-circ-OGDH#1 (si-circ-OGDH) with better knockdown efficiency was chose for follow-up analysis. MTT and colony formation assays exhibited that circ-OGDH silencing inhibited the proliferation of TE-1 and Eca-109 cells (Figure 2B and C). Cell apoptosis analysis manifested that the apoptotic rate was overtly elevated in si-circ-OGDH-transfected TE-1 and Eca-109 cells (Figure 2D). Transwell assay revered that circ-OGDH depletion significantly repressed cell migration and invasion in TE-1 and Eca-109 cells in (Figure 2E and F). Cell cycle analysis exhibited that circ-OGDH inhibition resulted in the arrest of TE-1 and Eca-109 cells in G0/G1 stage (Figure 2G and H). Moreover, circ-OGDH silencing reduced glutamine consumption, and α-KG production, and ATP content in TE-1 and Eca-109 cells (Figure 2I-K). In addition, circ-OGDH knockdown decreased the level of GLS1 protein in TE-1 and Eca-109 cells (Figure 2L). Together, these results indicated that circ-OGDH promoted proliferation, metastasis, invasion, and glutamine metabolism of ESCC cells in vitro.
Circ-OGDH Was Identified as a Sponge for miR-615-5p
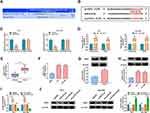
To explore the regulatory mechanism of circ-OGDH in ESCC, we sought for miRs that might interact with circ-OGDH. As displayed in Figure 3A, circ-OGDH might serve as a molecule sponge for miR-615-5p. Subsequently, we established a luciferase plasmid containing wt-circ-OGDH or mut-circ-OGDH to analyze the relationship between circ-OGDH and miR-615-5p (Figure 3B). Dual-luciferase reporter assay showed that the luciferase activity of the luciferase plasmid carrying wt-circ-OGDH was reduced in the presence of miR-615-5p mimic in TE-1 and Eca-109 cells, but the luciferase activity of the luciferase plasmid with mut-circ-OGDH did not change (Figure 3C). RIP assay exhibited that miR-615-5p and circ-OGDH were overtly enriched in RNA complexes in the Ago2 group compared with the IgG group (Figure 3D). We also observed significant downregulation of miR-615-5p in ESCC tissues and cells (Figure 3E and F). Also, circ-OGDH expression was significantly increased in TE-1 and Eca-109 cells transfected with circ-OGDH than that in the control group (Figure 3G). Additionally, miR-615-5p expression was elevated in TE-1 and Eca-109 cells with inhibition of circ-OGDH and decreased in TE-1 and Eca-109 cells with overexpression of circ-OGDH (Figure 3H). Collectively, these results manifested that circ-OGDH acted as a sponge for miR-615-5p in ESCC cells.
Circ-OGDH Regulated Proliferation, Metastasis, Invasion, and Glutamine Metabolism of ESCC Cells by Sponging miR-615-5p
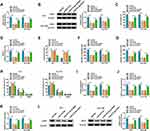
Subsequently, we conducted rescue experiments to further survey whether circ-OGDH regulated proliferation, metastasis, invasion, and glutamine metabolism of ESCC cells by adsorbing miR-615-5p. QRT-PCR exhibited that miR-615-5p inhibition reverted the upregulation of miR-615-5p in circ-OGDH-silenced TE-1 and Eca-109 cells (Figure 4A). As expected, miR-615-5p silencing reverted impacts of circ-OGDH depletion on proliferation and apoptosis of TE-1 and Eca-109 cells (Figure 4B-D). Also, miR-615-5p downregulation reversed the inhibitory influence of circ-OGDH knockdown on metastasis, invasion, and cell cycle progression of TE-1 and Eca-109 cells (Figure 4E-G). Moreover, miR-615-5p inhibition restored the decrease of glutamine consumption, α-KG production, and ATP content in circ-OGDH-silenced TE-1 and Eca-109 cells (Figure 4H-J). In addition, silence of miR-615-5p reversed the downregulation of GLS1 protein in TE-1 and Eca-109 cells induced by circ-OGDH knockdown (Figure 4K). In sum, these data suggested that circ-OGDH adsorbed miR-615-5p to regulate proliferation, metastasis, invasion, and glutamine metabolism of ESCC cells.
PDX1 Was a Target of miR-615-5p in ESCC Cells
To investigate the regulatory mechanism of miR-615-5p in ESCC, we searched the targets of miR-615-5p. As displayed in Figure 5A, miR-615-5p might target PDX1. Thus, we constructed a luciferase plasmid with wt-PDX1 3ʹUTR or mut-PDX1 3ʹUTR to further validate the relationship between miR-615-5p and PDX1 (Figure 5B). We observed that miR-615-5p overexpression repressed the luciferase activity in TE-1 and Eca-109 cells carrying the luciferase plasmid containing wt-PDX1 3ʹUTR but not the luciferase plasmid containing mut-PDX1 3ʹUTR (Figure 5C). Moreover, miR-615-5p and PDX1 were markedly co-enriched in the Ago2 group (Figure 5D). We also examined the levels of PDX1 mRNA and protein in ESCC tissues and cells and the results exhibited higher expression levels of PDX1 mRNA and protein in ESCC tissues and cells (Figure 5E-H). Additionally, miR-615-5p expression was reduced in TE-1 and Eca-109 cells transfected with miR-615-5p inhibitor and increased in TE-1 and Eca-109 cells transfected with miR-615-5p mimic (Figure 5I). As expected, miR-615-5p overexpression markedly decreased PDX1 protein level in TE-1 and Eca-109, but miR-615-5p inhibition had an opposite influence (Figure 5J). These data indicated that miR-615-5p directly targeted PDX1 in ESCC cells.
MiR-615-5p Regulated Proliferation, Metastasis, Invasion, and Glutamine Metabolism of ESCC Cells Through Targeting PDX1
To test whether the miR-615-5p/PDX1 axis regulated the proliferation, metastasis, invasion, and glutamine metabolism of ESCC cells, we performed rescue experiments. After PDX1 introduction, the downregulation of PDX1 mRNA and protein in miR-615-5p-overexpressed TE-1 and Eca-109 cells was partially overturned (Figure 6A and B). Moreover, miR-615-5p overexpression repressed proliferation and accelerated apoptosis of TE-1 and Eca-109 cells, but these effects were restored by forcing PDX1 expression (Figure 6C-E). Also, elevated PDX1 expression overturned the repressive effect of miR-615-5p mimic on migration, invasion, and cell cycle progression of TE-1 and Eca-109 cells (Figure 6F-H). As expected, miR-615-5p elevation reduced glutamine consumption, α-KG production, and ATP content in TE-1 and Eca-109 cells, but these declines were offset by PDX1 overexpression (Figure 6I-K). Additionally, the forcing expression of PDX1 reversed the downregulation of GLS1 protein in miR-615-5p-increased TE-1 and Eca-109 cells (Figure 6L). These results indicated that miR-615-5p repressed proliferation, metastasis, invasion, and glutamine metabolism of ESCC cells through targeting PDX1.
Circ-OGDH Sponged miR-615-5p and Increased PDX1 Expression
Based on the above findings, we further surveyed whether circ-OGDH functioned as a ceRNA (competing endogenous RNA) and regulated PDX1 expression by sponging miR-615-5p. The results showed that circ-OGDH inhibition reduced PDX1 protein level in TE-1 and Eca-109 cells, but this decline was reversed after miR-615-5p downregulation (Figure 7A and B). These data indicated that circ-OGDH functioned as a miR-615-5p sponge and regulated PDX1 expression via adsorbing miR-615-5p in ESCC cells.
Circ-OGDH Inhibition Reduced ESCC Growth in vivo
To validate the function of circ-OGDH in ESCC, we established ESCC animal models by injecting mice with TE-1 cells carrying sh-circ-OGDH or sh-NC. We observed that tumor volume and weight were overtly smaller and lighter in mice injected with TE-1 cells carrying with sh-circ-OGDH in contrast to the control group (Figure 8A-C). QRT-PCR analysis of tumor tissues revealed that circ-OGDH and PDX1 mRNA expression were decreased mice tumor tissues in the sh-circ-OGDH group, whereas miR-615-5p expression was elevated (Figure 8D). Additionally, PDX1 protein level was also downregulated in mice tumor tissues in the sh-circ-OGDH group (Figure 8E). In sum, these data suggested that circ-OGDH accelerated ESCC growth in vivo.
Discussion
Emerging studies have discovered that circRNAs have great potential to become therapeutic targets for cancer.16 At present, many circRNAs have been uncovered to be implicated in the tumorigenesis of ESCC. It was reported that circRNA circ-GSK3β,17 circRNA circ-LPAR3,18 and circRNA circ-NTRK2 played a tumor-promoting role in ESCC, while circRNA circ-VRK1,19 circRNA circ-SMAD7,20 and circRNA circ-FOXO321 acted as a tumor suppressor. Even so, the exact mechanism of circRNAs in ESCC is still unknown.
Glutamine metabolism has been demonstrated to be indispensable for the development of tumors.22 Also, the disruption of glutaminolysis offers a promising approach for the treatment of ESCC.23 GLS1, which converts glutamine to glutamate, is the first enzyme in the glutamine decomposition pathway.24 OGDH, the host gene of circ-OGDH, plays a regulatory role at the intersection of the TCA cycle and glutamine metabolism.25,26 OGDH played a promoting effect in tumor progression via regulation of mitochondrial bioenergetics and Wnt/beta-catenin pathway in gastric cancer.26 Our data indicated that circ-OGDH silencing decreased GLS1 protein level, glutamine consumption, α-KG production, and ATP content in ESCC cells, illustrating circ-OGDH facilitated glutamine metabolism in ESCC cell. Furthermore, circ-OGDH knockdown repressed ESCC cell growth in vivo and constrained ESCC cell proliferation, metastasis, and invasion in vitro, indicating that circ-OGDH played a promoting influence on tumor growth in ESCC. Hou et al disclosed that circ-OGDH functioned as miR-564 decoy and promoted cell invasion and proliferation by increasing TPX2 expression through adsorbing miR-564 in ESCC.15 These data indicated that circ-OGDH elevated glutamine metabolism and accelerated tumor growth in ESCC.
The ceRNA hypothesis indicates that circRNAs can absorb miRs through miR response elements, thus the downstream targets of mIRs.27 Based on this hypothesis, we proved that circ-OGDH served as a miR-615-5p molecule sponge by bioinformatics analysis, dual-luciferase reporter, and RIP assay. MiR-615-5p had been unveiled to be downregulated in many tumors,28–31 and its overexpression exerted an inhibitory effect on tumor growth. For instance, circRNA circ-PUM1 sponged miR-615-5p to accelerate the tumorigenesis of ovarian cancer.31 MiR-615-5p repressed pancreatic ductal adenocarcinoma progression via targeting AKT2.32 Also, low miR-615-5p expression was associated with advanced tumor-node-metastasis stage in ESCC, and miR-615-5p mimic curbed ESCC cell invasion and migration in vitro.33 Our results also indicated that miR-615-5p expression was reduced in ESCC. Moreover, miR-615-5p silencing reversed the repressive impact of circ-OGDH knockdown on glutamine metabolism and malignant behaviors of ESCC cells. Accordingly, we inferred that circ-OGDH increased glutamine metabolism and accelerated malignant behaviors of ESCC cells through sponging miR-615-5p.
PDX1, located at chromosome 13q12.1, is an essential transcription factor for pancreatic development.34 PDX1 could resist the formation of pancreatic ductal adenocarcinoma derived from pancreatic intraepithelial neoplasia, but after tumor transformation, the function of PDX1 changed from suppressing-tumor to carcinogenic.35 Previous study had uncovered that lncRNA MAFG-AS1 facilitated aerobic glycolysis, invasion, and migration of ESCC cells through adsorbing miR-765 and increasing PDX1 expression.36 Herein, PDX1 was also highly expressed in ESCC. Moreover, PDX1 was validated as a target of miR-615-5p, and PDX1 elevation restored the repressive effect of miR-615-5p mimic on glutamine metabolism and accelerated malignant behaviors of ESCC cells. Hence, we concluded that circ-OGDH acted as a ceRNA and accelerated glutamine metabolism and tumor growth through upregulating PDX1 via sponging miR-615-5p in ESCC.
In sum, circ-OGDH exerted a cancerogenic role in ESCC. Mechanically, circ-OGDH sponged miR-615-5p to release PDX1, thereby elevating glutamine metabolism and facilitating tumor growth in ESCC. The study revealed that circ-OGDH was a promising target for ESCC treatment.
Funding
This study was supported by grant from Chengde City Science and Technology Research and Development Project 2016 (No: 201606A036).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. doi:10.1053/j.gastro.2017.08.023
3. Saeki H, Sohda M, Sakai M, Sano A, Shirabe K. Role of surgery in multidisciplinary treatment strategies for locally advanced esophageal squamous cell carcinoma. Ann Gastroenterol Surg. 2020;4(5):490–497. doi:10.1002/ags3.12364
4. Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet (London, England). 2017;390(10110):2383–2396. doi:10.1016/S0140-6736(17)31462-9
5. Guo XF, Mao T, Gu ZT, Ji CY, Fang WT, Chen WH. Clinical study on postoperative recurrence in patients with pN0 esophageal squamous cell carcinoma. J Cardiothorac Surg. 2014;9:150. doi:10.1186/s13019-014-0150-4
6. Li T, Le A. Glutamine metabolism in cancer. Adv Exp Med Biol. 2018;1063:13–32.
7. Xiao D, Zeng L, Yao K, Kong X, Wu G, Yin Y. The glutamine-alpha-ketoglutarate (AKG) metabolism and its nutritional implications. Amino Acids. 2016;48(9):2067–2080. doi:10.1007/s00726-016-2254-8
8. Matés JM, Campos-Sandoval JA, Santos-Jiménez JDL, Márquez J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett. 2019;467:29–39. doi:10.1016/j.canlet.2019.09.011
9. Jin L, Alesi GN, Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 2016;35(28):3619–3625. doi:10.1038/onc.2015.447
10. Zhang P, Chao Z, Zhang R, et al. Circular RNA regulation of myogenesis. Cells. 2019;8(8):885. doi:10.3390/cells8080885
11. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
12. Zhou MY, Yang JM, Xiong XD. The emerging landscape of circular RNA in cardiovascular diseases. J Mol Cell Cardiol. 2018;122:134–139. doi:10.1016/j.yjmcc.2018.08.012
13. Ding L, Zhao Y, Dang S, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45. doi:10.1186/s12943-019-1006-2
14. Wang S, Zhang Y, Cai Q, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer. 2019;18(1):145. doi:10.1186/s12943-019-1078-z
15. Hou Y, Liu H, Pan W. Knockdown of circ_0003340 induces cell apoptosis, inhibits invasion and proliferation through miR-564/TPX2 in esophageal cancer cells. Exp Cell Res. 2020;394(2):112142. doi:10.1016/j.yexcr.2020.112142
16. Ma Z, Shuai Y, Gao X, Wen X, Circular JJ. RNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):8. doi:10.1186/s12943-019-1113-0
17. Hu X, Wu D, He X, et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol Cancer. 2019;18(1):160. doi:10.1186/s12943-019-1095-y
18. Shi Y, Fang N, Li Y, et al. Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111(8):2824–2836. doi:10.1111/cas.14511
19. He Y, Mingyan E, Wang C, Liu G, Shi M, Liu S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol. 2019;125:116–123. doi:10.1016/j.ijbiomac.2018.11.273
20. Zhang Y, Wang Q, Zhu D, Rong J, Shi W, Cao X. Up-regulation of circ-SMAD7 inhibits tumor proliferation and migration in esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;111:596–601. doi:10.1016/j.biopha.2018.12.116
21. Xing Y, Zha WJ, Li XM, et al. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem. 2020;121(3):2595–2605. doi:10.1002/jcb.29481
22. Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36(10):1302–1315. doi:10.15252/embj.201696151
23. Qie S, Yoshida A, Parnham S, et al. Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma. Nat Commun. 2019;10(1):1296. doi:10.1038/s41467-019-09179-w
24. Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30(7):671–678. doi:10.1038/nbt.2285
25. Denton RM, Pullen TJ, Armstrong CT, Heesom KJ, Rutter GA. Calcium-insensitive splice variants of mammalian E1 subunit of 2-oxoglutarate dehydrogenase complex with tissue-specific patterns of expression. Biochem J. 2016;473(9):1165–1178. doi:10.1042/BCJ20160135
26. Lu X, Wu N, Yang W, Sun J, Yan K, Wu J. OGDH promotes the progression of gastric cancer by regulating mitochondrial bioenergetics and Wnt/β-catenin signal pathway. Onco Targets Ther. 2019;12:7489–7500. doi:10.2147/OTT.S208848
27. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
28. El Tayebi HM, Hosny KA, Esmat G, Breuhahn K, Abdelaziz AI. miR-615-5p is restrictedly expressed in cirrhotic and cancerous liver tissues and its overexpression alleviates the tumorigenic effects in hepatocellular carcinoma. FEBS Lett. 2012;586(19):3309–3316. doi:10.1016/j.febslet.2012.06.054
29. Gao W, Gu Y, Li Z, et al. miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene. 2015;34(13):1629–1640. doi:10.1038/onc.2014.101
30. Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):13. doi:10.1186/s12943-019-0943-0
31. Guan X, Zong ZH, Liu Y, Chen S, Wang LL, Zhao Y. circPUM1 promotes tumorigenesis and progression of ovarian cancer by Sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892. doi:10.1016/j.omtn.2019.09.032
32. Sun Y, Zhang T, Wang C, et al. MiRNA-615-5p functions as a tumor suppressor in pancreatic ductal adenocarcinoma by targeting AKT2. PLoS One. 2015;10(4):e0119783. doi:10.1371/journal.pone.0119783
33. Yang B, Xie R, Wu SN, Gao CC, Yang XZ, Zhou JF. MicroRNA-615-5p targets insulin-like growth factor 2 and exerts tumor-suppressing functions in human esophageal squamous cell carcinoma. Oncol Rep. 2018;39(1):255–263. doi:10.3892/or.2017.6079
34. Jennings RE, Berry AA, Strutt JP, Gerrard DT, Hanley NA. Human pancreas development. Development (Cambridge, England). 2015;142(18):3126–3137. doi:10.1242/dev.120063
35. Roy N, Takeuchi KK, Ruggeri JM, et al. PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes Dev. 2016;30(24):2669–2683. doi:10.1101/gad.291021.116
36. Qian CJ, Xu ZR, Chen LY, Wang YC, Yao J. LncRNA MAFG-AS1 accelerates cell migration, invasion and aerobic glycolysis of esophageal squamous cell carcinoma cells via miR-765/PDX1 axis. Cancer Manag Res. 2020;12:6895–6908. doi:10.2147/CMAR.S262075
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.