Back to Journals » OncoTargets and Therapy » Volume 14
CircPTCH1 Promotes Migration in Lung Cancer by Regulating MYCN Expression Through miR-34c-5p
Received 9 June 2021
Accepted for publication 17 August 2021
Published 10 September 2021 Volume 2021:14 Pages 4779—4789
DOI https://doi.org/10.2147/OTT.S324015
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
ZhenYu Shen, ShengHua Sun
Pulmonology and Critical Care Medicine Department, The Third Xiangya Hospital of Central South University, Changsha, 410013, People’s Republic of China
Correspondence: ShengHua Sun
Pulmonology and Critical Care Medicine Department, The Third Xiangya Hospital of Central South University, Changsha, 410013, People’s Republic of China
Tel/Fax +86 731-88618576
Email [email protected]
Background: The incidence rate and mortality rate of lung cancer are the highest in the world. Therefore, further studies are needed to reveal the molecular mechanism of lung cancer progression and development. Previous study demonstrated that the deregulation of circRNAs can regulate cell biological functions in tumorigenesis and development. However, the roles of circPTCH1 in lung cancer have not yet been revealed.
Materials and Methods: The expression levels of circPTCH1, miR-34c-5p, and MYCN were measured by RT-PCR in lung cancer tissues and cells; dual-luciferase reporter and RIP assay showed that circRNA served as a sponge for miRNA, and miRNA could target mRNA. In vitro, effects of si-circPTCH1 can regulate lung cancer cells’ migration, invasion were detected by CCK-8 assay, wound healing assay, and transwell assay.
Results: Our research demonstrated that the expression of circPTCH1 was upregulated in lung cancer tissues and cell lines and increased in metastatic tissues compared to that of non-metastatic tissues. circPTCH1 sponging miR-34c-5p to target MYCN was revealed by dual-luciferase reporter and a RIP assay. In addition, the expression level of miR-34c-5p was reduced in lung cancer tumor tissues, and MYCN was significantly increased in lung cancer tumor tissues. Pearson correlation analysis showed that miR-34c-5p with circPTCH1 and MYCN had a moderately negative correlation, and there was a moderately positive correlation between circPTCH1 and MYCN. Further, cytological studies found that circPTCH1 reduced lung cancer cells’ migration and invasion by targeting MYCN via miR-34c-5p.
Conclusion: circPTCH1 plays a tumor enhancement role in lung cancer and that can effectively promote migration, invasion and EMT by targeting the miR-34c-5p/MYCN axis. circPTCH1 may be a novel potential treatment and diagnosis biomarker for lung cancer.
Keywords: lung cancer, circPTCH1, biomarkers, miR-34c-5p, MYCN
Introduction
The incidence and mortality rates of lung cancer are the highest in the world. More than one million people die of lung cancer each year1 Lung cancer are divided into small cell lung cancer (SCLC) and non-small cell lung cancer (non-small cell lung cancer). There are many differences between clinicopathological features and prognosis them the 5-year overall survival rate of lung cancer is only about 18%. In addition, due to lack of relatively specific tumor markers for the diagnosis and treatment of lung cancer, it also has increased the challenge for the diagnosis and treatment of lung cancer.2,3 Therefore, it is necessary to study the molecular mechanism of lung cancer and explore the potential biomarkers for lung cancer.
At present, the discovery of circRNA has become a research hotspot in the field of non-coding RNA, and the focus of function and mechanism has become a hotspot in the current research. As a special non-coding RNA, circRNA is a new kind of RNA. CircRNA is different from linear RNA and has a closed loop structure, which is stable expression, not easy to degrade and affected by RNA exonuclease. CircRNAs have miRNA binding sites and act as miRNA sponges, which can block or reduce miRNA expression and promote the expression of target mRNA;4 at present, studies suggest that circRNA may be a new tumor molecular marker.5–7 However, the role and mechanism of circRNA in tumorigenesis and development are rarely studied.
More and more studies have shown that non-coding RNA plays a key role in a variety of human diseases, such as circRNA can directly participate in the regulation of cell proliferation, differentiation, migration and invasion by activating a variety of signaling pathways, thus playing an important role in the occurrence and development of malignant tumors.8–13 and a variety of circular RNAs play an important regulatory role in the proliferation, invasion and metastasis of lung cancer cells.14–16 Studies have shown that circPTCH1 is related to the occurrence and development of tumor, participates in the formation, invasion and metastasis of tumor, and may play the role of oncogene in tumor,17 but the mechanism of circPTCH1 in lung cancer is still unclear. Therefore, this study examined the expression level of circPTCH1 in 35 cases of lung cancer and corresponding normal lung tissues, and found that the expression level of circPTCH1 in lung cancer tissues was significantly higher than that in normal lung tissues, suggesting that circPTCH1 plays an important role in the occurrence and development of lung cancer, but its exact mechanism needs to be further explored.
Materials and Methods
Bioinformatics Analysis
We used online databases (circinteractome, starbase and circbank) to find the potential miRNA of circPTCH1, and starbase databases were used to predict the target genes of miR-34c-5p.
Clinical Specimens
Thirty-five pairs of lung cancer clinical specimens and adjacent normal tumors were collected from the Third Xiangya Hospital of Central South University. And the adjacent normal tumor tissues belong to the normal tissues, which have no cancer cell invasion. None of them had undergone chemotherapy, radiotherapy or any anti-cancer treatment before surgery, and all the clinical specimens were immediately flash-frozen in liquid nitrogen until RNA extraction. This study was performed referring to the Helsinki Declaration and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University. All lung cancer patients provided written informed consent and the study abided by the right to privacy of human rights subjects.
Cell Culture and Cell Transfection
BEAS-2B, A549, NCI-H1299, SK-MES-1, and NCI-H661 were obtained from the Cell Collection Committee of the Chinese Academy of Sciences (Shanghai, China). Cells were incubated by DMEM medium (Gibco, USA) with 10% FBS (Gibco, Australia) at 37°C, 5%CO2. The cells line are authenticated by STR method (Biowing, China). circPTCH1 si-RNA and circPTCH1 over-expression plasmids were synthesized by GenePharma (Shanghai, China). MiRNAs inhibitors were purchased from RiboBio (Guangzhou, China) and cell transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol.
RNA Isolation, RNase R Treatment and Real-Time PCR
The total RNA was extracted from 35 pairs of lung cancer clinical specimens and adjacent normal specimens or lung cancer cells using TRIzol reagent (Invitrogen, CA, USA). 10 μg total RNA isolation was separated by 2 U/μg RNase R (Epicentre Technologies) for 30 min at 37°C. Random primers and stem-loop primers were used to synthesize cDNA using the TaKaRa system (Takara, Dalian, China). RT-PCR primers were purchased from RiboBio. circPTCH1-Forward: ACCAAAAGCCAAGGCAG -CGG, circPTCH1-Reverse: CCTCGCGTCGATATAAATCC; miR-34c-5p-Forward: CGGAGGCAGTGTAGTTAGCT, miR-34c-5p-Reverse: GTGCAGGGTCCGAGGT; U6-Forward:CTCGCTTCGGCAGCACA, U6-Reverse:AACGCTTCACGAATTTGC -GT; MYCN-Forward: ACCCGGACGAAGATGACTTCT, MYCN-Reverse: CAGCTCGTTCTCAAGCAGCAT; GAPDH-Forward: ACAACTTTGGTATCGTG GAAGG, GAPDH-Reverse: GCCATCACGCCACAGTTTC; Real-time PCR was detected by the CFX96 Tm Real-Time System (Bio-Rad, USA). The calculation method of relative expression was using the comparative Ct(2−ΔΔCt) method.
Wound Healing Assay
1×106 cells/well A549 or SK-MES-1 was seeded into 6-well plates. When the cells reached 90% confluency, the tip of 200 μL pipette was used to scratch the cell monolayer, and then a fresh serous medium was used to wash the plates three times. After 24 h, the wound width was calculated using Image J software.
Transwell Assay
1×104 cells/well A549 or SK-MES-1 cells were seeded on the upper chambers with 2% serum medium and medium containing 10% serum medium were added to the lower chambers. Then, the cells were incubated at 37°C for 24 h. The number of migrating cells was counted to calculate the average number of migrated cells per plate.
Dual-Luciferase Reporter Assay
Cells were cotransfected with circPTCH1-miR-34c-5p and MYCN-miR-34c-5p into the luciferase gene (wild-type or mutant-type), and the specific operation was carried out according to the manufacturer’s protocol. After 48 h, luciferase activities were calculated for each well using the Dual Luciferase Reporter Assay System (Promega).
RNA-Binding Protein Immunoprecipitation (RIP)
RIP assay (Millipore, Billerica, MA) was carried out according to the manufacturer’s instructions. The RIP lysate was obtained and centrifuged at 12,000 rpm for 10 minutes. Add 100 µL of the supernatant to the RIP immunoprecipitation buffer containing the magnetic bead-antibody complex; then, the RNA was purified and obtained from TRIzol. Finally, the immunoprecipitated RNA was analyzed by RT-PCR.
Tumor Xenografts
2×106 cells were stably transfected with si‐circPTCH1 plasmids or circPTCH1 overexpression plasmids or negative control vector were injected into the upper back of nude mice. Tumor weight was detected every 7 days. The volume of tumors was calculated by using the following formula: V = 0.5 × L × W2. One month later, all animals were sacrificed, and the tumor volume was recorded. The laboratory animals were approved by the medical laboratory animal ethics committee of the Third Xiangya Hospital of Central South University. Instructive notions with respect to caring for laboratory animals (which is released by the Ministry of Science and Technology of the People’s Republic of China in September 30th, 2006) were followed for the welfare of the animals.
Western Blot Assay
Proteins were extracted from cells using RIPA with proteinase inhibitors (Sigma-Aldrich, USA) and the concentrations of proteins were measured using BCA Protein Assay kits (Thermo Scientific, MA). After separation by 10% SDS/PAGE, proteins were blocked with 3% non-fat milk for 1 h and immunoblotted with the primary antibodies at 4°C overnight. Proteins were incubated with secondary antibodies for 1 h. After TBST washing 3 times and the samples were measured using the chemiluminescence image system (Bio-Rad, USA).
Statistic Analysis
All data were analyzed by SPSS 20.0, P < 0.05 indicated statistical significant. Data represented as means ± standard, and Student’s t-test for two groups and one-way ANOVA with post hoc Bonferroni test for three or more groups were used to assess the statistical significance. The Pearson correlation between miR-34c-5p with circPTCH1 and MYCN was calculated to evaluate the reverse expression pattern.
Results
circPTCH1 is Up-Regulated in Lung Cancer Patients and Cell Lines
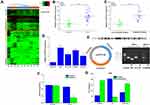
High throughput sequencing was used to detect the expression of circrna in five lung cancer tissues and five matched lung tissues. In total, 253 circRNAs were considered to be significantly differentially expressed (up-regulated 101 circRNAs, down-regulated 142 circRNAs). The significantly dysregulated circRNAs were presented in a clustered heat map (P<0.001, Figure 1A). Among them, CircPTCH1 was the most significantly overexpressed circRNA in lung cancer tissues; then, RT-PCR results showed that the expression of circPTCH1 was higher in lung cancer tissues compared to that in adjacent normal tumors (P<0.001, Figure 1B) and significantly increased in metastatic tissues compared to that of non-metastatic tissues (P<0.001, Figure 1C, Supplementary Table S1). Besides, the expression of circPTCH1 was significantly increased in A549, NCI-H1299, SK-MES-1, and NCI-H661 compared with BEAS-2B (Figure 1D). The convergent primers and divergent primers were used to amplify linear or circRNA PTCH1 by cDNA and genomic DNA (gDNA). RT-PCR results showed that divergent primers could amplify by cDNA but could not amplify by gDNA (Figure 1E). Then, the linear mRNA PTCH1 expression was significantly decreased, but circPTCH1 expression did not have any significant change after the RNase R treatment (Figure 1F). To investigate the subcellular localization of circPTCH1, we detected the distribution of circPTCH1 in the nuclear and cytoplasmic fractions of A549 cells, and FISH result found that circPTCH1 was predominately distributed in the cytoplasm (Figure 1G and H; Figure 3J).
circPTCH1 Regulates Lung Cancer Cells Migration, and Invasion
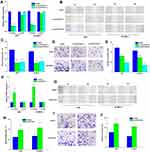
QRT-PCR assay that showed si-circPTCH1 could significantly decreased the expression of circPTCH1 in A549 and SK-MES-1 cells, and there was no significant change in PTCH1 mRNA expression (Figure 2A). Wound healing assay and transwell assay revealed that circPTCH1 knockdown group could suppress A549 and SK-MES-1 cells’ invasion and migration (Figure 2B–E). RT-PCR assay showed that overexpression of circPTCH1 plasmid could significantly increase the expression of circPTCH1 and not significantly effect mRNA PTCH1 level in A549 and SK-MES-1 cells (Figure 2F), and wound healing assay and transwell assay revealed that overexpression of circPTCH1 could promote A549 and SK-MES-1 cells’ invasion and migration (Figure 2G–J).
circPTCH1 Can Binding to miR-34c-5p, and MYCN is a Direct Target of miR-34c-5p in Lung Cancer
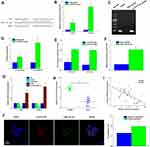
We used three publicly online tools: circinteractome (https://circinteractome.nia.nih.gov/), starbaseV3.0 (http://starbase.sysu.edu.cn/) and circbank (http://www.circbank. cn/index.html) to predict the possible binding miRNAs of circPTCH1. We compared three online databases, and 2 potential miRNAs were selected (Figure 3A). A biotin-linked circPTCH1 probe was designed to pull down circPTCH1 in A549 cells, the pull-down effectiveness was obviously elevated in cells overexpressing circPTCH1circPTCH1 probe compared with the NC probe, and miR-34c-5p was the most enriched (Figure 3B–D). Dual-luciferase reporter assay revealed that 2 miRNAs including miR-34c-5p, miR-34b-5p were significantly pulled down by circPTCH1 in A549 cells (Figure 3E). In addition, circPTCH1 was abundantly pulled down by the miR-34c-5p probe compared with the NC probe (Figure 3F). RT-PCR assay showed that the expression levels of miR-34c-5p was up-regulated when transfected with si-circPTCH1 plasmid and decreased by circPTCH1 over-expression plasmid in A549 and SK-MES-1 cells (Figure 3G). QRT-PCR analysis of the expression levels of miR-34c-5p was decreased in lung cancer tissues compared with normal tissues. Meanwhile, between circPTCH1 and miR-34c-5p expression has observed a negative correlation with Pearson correlation in lung cancer patients (Figure 3H and I). FISH assay revealed that circPTCH1 and miR-34c-5p were colocalized in the cytoplasm (Figure 3J).
We used starbase V3.0 databases (http://starbase.sysu.edu.cn/) to predict the target genes of miR-34c-5p and MYCN as a direct target of miR-34c-5p. Dual-luciferase reporter assay confirmed that miR-34c-5p considerably decreased the luciferase activity in the MYCN-WT group, but not in the MYCN-MUT group when compared with the control group (Figure 4A and E). The expression of MYCN was significantly increased in A549, NCI-H1299, SK-MES-1, and NCI-H661 compared with BEAS-2B, and also increased in lung cancer tissues compared with normal tissues (Figure 4B and C). Meanwhile, between MYCN and miR-34c-5p expression has observed a negative correlation with Pearson correlation in lung cancer patients (Figure 4D). Furthermore, the expression of MYCN was significantly decreased by miR-34c-5p mimic group and restored by miR-34c-5p mimic+MYCN (Figure 4F). Wound healing assay and transwell assay revealed that miR-34c-5p mimic group could also suppress invasion and migration, and the function was restored by miR-34c-5p mimic+MYCN (Figure 4G and H).
circPTCH1 Knock Down Inhibits Lung Cancer Cells Migration, Invasion and EMT by Targeting MYCN via miR-34c-5p
The study also investigated whether or not circPTCH1 could directly target MYCN via miR-34c-5p to induce EMT in lung cancer cells. Our results showed that the expression of MYCN was significantly decreased by si-circPTCH1 group and recovered by si-circPTCH1+miR-34c-5p inhibitor (Figure 5A). On the contrary, the expression of MYCN was significantly increased by circPTCH1 overexpression group and recovered by circPTCH1+miR-34c-5p mimic (Figure 5B). Wound healing assay and transwell assay revealed that si-circPTCH1 group could also suppress invasion and migration, while the function was restored by si-circPTCH1+miR-34c-5p inhibitor group (Figure 5C and D). WB results showed that the expression of E-cadherin protein was significantly increased si-circPTCH1 and rescued by circPTCH1+miR-34c-5p mimic. However, MYCN and Vimentin were decreased by si-circPTCH1 and rescued by circPTCH1+miR-34c-5p mimic (Figure 5E and F). BALB/c nude mice experiments obtained similar results (Figure 5G). The protein expression of E-cadherin, MYCN, and Vimentin was significantly decreased with si-circPTCH1 group by immunofluorescence.
Discussion
Non-small cell lung cancer is the most common type of lung cancer, early symptoms are not obvious, difficult to diagnose, lack of effective treatment, and high mortality.18,19 Therefore, it is urgent to further explore the pathogenesis and provide sensitive, specific and effective indicators for the diagnosis and treatment of lung cancer. The aim of this study was to investigate the effects of circPTCH1 on migration and invasion of lung cancer cells, so as to analyze the biological function of circPTCH1 in lung cancer cells. And to explore the regulatory mechanism of circPTCH1 in the occurrence and development of lung cancer, so as to provide a new target for lung cancer treatment.
MicroRNA is a kind of small non-coding RNA, which binds to mRNA through base complementary pairing, so as to inhibit miRNA translation. Several different studies have found that miRNAs participate in the regulation of a variety of cell functions, and many miRNAs have abnormal expression in cancer,20,21,22 indicating that different miRNAs play an important regulatory role in cancer and have great application value in clinical diagnosis, prognosis evaluation and treatment. Many studies have shown that miR-34c-5p is abnormally low expressed in many tumors, such as colorectal cancer, gastric cancer, lung cancer, prostate cancer and so on,23,24–26 and plays an anti-cancer role in tumor development. The down-regulation of miR-34c-5p is closely related to the biological functions of a variety of tumors, such as proliferation, cell cycle progression, metastasis, invasion and chemoradiotherapy resistance.27,28 Many studies have shown that miR-34c-5p directly targets many genes and has different target genes in different tumors, such as E2F3, Bcl-2, c-Met and so on.29,30–32 Therefore, miR-34c-5p directly targeted gene in tumor remains to be further elucidated.
The N-myc protein encoded by MYCN gene is an important transcription factor, which can regulate the expression of a variety of target genes and thus participate in a variety of cell biological functions and signaling pathways.33,34 Studies have shown that MYCN can directly target cell cycle-related genes, reduce G1 phase arrest, promote cell into S phase and promote cell proliferation.33,35 The abnormal expression of MYCN was found in neurogenic tumors, including neuroblastoma, neuroendocrine prostate cancer and small cell lung cancer.36–40
Conclusion
In conclusion, this study demonstrated that circPTCH1 is more highly expression in lung cancer tissues and cell lines, and circPTCH1 acts as a sponge for miR-34c-5p to regulate MYCN. Moreover, circPTCH1 inhibits migration of lung adenocarcinoma by regulating mir-34c-5p to target MYCN gene, which provides a new theoretical basis for the prognosis and treatment of lung cancer.
Author Contributions
ZhenYu Shen performed the experiments, analysed the data and wrote the paper. ZhenYu Shen1, ShengHua Sun conceptualized the study design, and contributed to data analysis and experimental materials. All authors read and approved the final manuscript. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval for the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report that they have no competing interests in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Sun J, Xie T, Jamal M, et al. CLEC3B as a potential diagnostic and prognostic biomarker in lung cancer and association with the immune microenvironment. Cancer Cell Int. 2020;20:106. doi:10.1186/s12935-020-01183-1
3. Ma C, Yang X, Xing W, et al. Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac Cancer. 2020;11(3):588–593. doi:10.1111/1759-7714.13300
4. Du YX, Guo LX, Pan HS, et al. Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion. Hum Cell. 2020;33(1):252–260. doi:10.1007/s13577-019-00298-6
5. Aishanjiang K, Wei XD, Fu Y, et al. Circular RNAs and hepatocellular carcinoma: new epigenetic players with diagnostic and prognostic roles. Front Oncol. 2021;11:653717. doi:10.3389/fonc.2021.653717
6. Li Y, Feng W, Kong M, et al. Exosomal circRNAs: a new star in cancer. Life Sci. 2021;269:119039. doi:10.1016/j.lfs.2021.119039
7. Fu L, Tang D, Sun A, et al. Recent progress in study of circRNAs and its role in leukemia. J Leukoc Biol. 2021;109(4):731–739. doi:10.1002/JLB.2RU0619-213R
8. Xu Z, Xiang W, Chen W, et al. Circ-IGF1R inhibits cell invasion and migration in non-small cell lung cancer. Thorac Cancer. 2020;11(4):875–887. doi:10.1111/1759-7714.13329
9. Zhang CC, Li Y, Feng XZ, et al. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered. 2021;12(1):414–425.
10. Gao W, Guo H, Niu M, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer. 2020;19(1):166. doi:10.1186/s12943-020-01279-2
11. Shang A, Gu C, Wang W, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol Cancer. 2020;19(1):117. doi:10.1186/s12943-020-01235-0
12. Jiao J, Zhang T, Jiao X, et al. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J Cell Physiol. 2020;235(2):1287–1295. doi:10.1002/jcp.29045
13. Bhat AA, Younes SN, Raza SS, et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol Cancer. 2020;19(1):57.
14. Wang C, Fen Y, Li B, et al. CircRNAsin lung-intestinal axis cancer. Curr Mol Med. 2020;11:2479.
15. Gao P, Wang Z, Hu Z, et al. Circular RNA circ_0074027 indicates a poor prognosis for NSCLC patients and modulates cell proliferation, apoptosis, and invasion via miR-185-3p mediated BRD4/MADD activation. J Cell Biochem. 2020;121(3):2632–2642. doi:10.1002/jcb.29484
16. Ashrafizadeh M, Zarrabi A, Hushmandi K, et al. Lung cancer cells and their sensitivity/resistance to cisplatin chemotherapy: role of microRNAs and upstream mediators. Cell Signal. 2021;78:109871.
17. Liu H, Hu G, Wang Z, et al. circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis. Theranostics. 2020;10(23):10791–10807. doi:10.7150/thno.47239
18. Lu Y, Ma T, Wang L, et al. [Advances in lymph node metastasis and lymph node dissection in early non-small cell lung cancer]. Zhongguo Fei Ai Za Zhi. 2019;22(8):520–525. [Portuguese]
19. Mott TF. Lung cancer: management. FP Essent. 2018;464:27–30.
20. Huang W, Yan Y, Liu Y, et al. Exosomes with low miR-34c-3p expression promote invasion and migration of non-small cell lung cancer by upregulating integrin alpha2beta1. Signal Transduct Target Ther. 2020;5(1):39. doi:10.1038/s41392-020-0133-y
21. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. doi:10.1038/s41568-020-00306-0
22. Ma LL, Liang L, Zhou D, et al. Tumor suppressor miR-424-5p abrogates ferroptosis in ovarian cancer through targeting ACSL4. Neoplasma. 2021;68(1):165–173. doi:10.4149/neo_2020_200707N705
23. Wu Z, Wu Y, Tian Y, et al. Differential effects of miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and invasion of glioma cells. Oncol Lett. 2013;6(5):1447–1452. doi:10.3892/ol.2013.1579
24. Luo Y, Wang D, Chen S, et al. The role of miR-34c-5p/Notch in epithelial-mesenchymal transition (EMT) in endometriosis. Cell Signal. 2020;72:109666. doi:10.1016/j.cellsig.2020.109666
25. Wang Y, Wang X, Tang J, et al. The study of mechanism of miR-34c-5p targeting FLOT2 to regulate proliferation, migration and invasion of osteosarcoma cells. Artif Cells Nanomed Biotechnol. 2019;47(1):3559–3568. doi:10.1080/21691401.2019.1640714
26. Xu X, Yan H, Zhang L, et al. Up-regulation of miR-34c-5p inhibits nasopharyngeal carcinoma cells by mediating NOTCH1. Biosci Rep. 2020;40:6. doi:10.1042/BSR20200302
27. Fereshteh Z, Schmidt SA, Al-Dossary AA, et al. Murine Oviductosomes (OVS) microRNA profiling during the estrous cycle: delivery of OVS-borne microRNAs to sperm where miR-34c-5p localizes at the centrosome. Sci Rep. 2018;8(1):16094. doi:10.1038/s41598-018-34409-4
28. Wan Y, Shen A, Qi F, et al. Pien Tze Huang inhibits the proliferation of colorectal cancer cells by increasing the expression of miR-34c-5p. Exp Ther Med. 2017;14(4):3901–3907. doi:10.3892/etm.2017.4972
29. Li F, Chen H, Huang Y, et al. miR-34c plays a role of tumor suppressor in HEC1-B cells by targeting E2F3 protein. Oncol Rep. 2015;33(6):3069–3074. doi:10.3892/or.2015.3894
30. Tu Y, Hu Y. MiRNA-34c-5p protects against cerebral ischemia/reperfusion injury: involvement of anti-apoptotic and anti-inflammatory activities. Metab Brain Dis. 2021;36(6):1341–1351. doi:10.1007/s11011-021-00724-5
31. Liu B, Gan W, Jin Z, et al. The role of miR-34c-5p in osteogenic differentiation of bone marrow mesenchymal stem cells. Int J Stem Cells. 2021. doi:10.15283/ijsc20188
32. Cai H, Yu Y, Ni X, et al. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 2020;11(12):1032. doi:10.1038/s41419-020-03247-6
33. Kong D, Fan S, Sun L, et al. Growth inhibition and suppression of the mTOR and Wnt/beta-catenin pathways in T-acute lymphoblastic leukemia by rapamycin and MYCN depletion. Hematol Oncol. 2021;39(2):222–230.
34. Lu Y, Yang Q, Su Y, et al. MYCN mediates TFRC-dependent ferroptosis and reveals vulnerabilities in neuroblastoma. Cell Death Dis. 2021;12(6):511. doi:10.1038/s41419-021-03790-w
35. Wu X, Nelson M, Basu M, et al. MYC oncogene is associated with suppression of tumor immunity and targeting Myc induces tumor cell immunogenicity for therapeutic whole cell vaccination. J Immunother Cancer. 2021;9(3). doi:10.1136/jitc-2020-001388.
36. Irtan S, Brisse HJ, Minard-Colin V, et al. Image-defined risk factor assessment of neurogenic tumors after neoadjuvant chemotherapy is useful for predicting intra-operative risk factors and the completeness of resection. Pediatr Blood Cancer. 2015;62(9):1543–1549. doi:10.1002/pbc.25511
37. Lee JK, Phillips JW, Smith BA, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29(4):536–547. doi:10.1016/j.ccell.2016.03.001
38. Berger A, Brady NJ, Bareja R, et al. N-Myc-mediated epigenetic reprogramming drives lineage plasticity in advanced prostate cancer. J Clin Invest. 2019;129(9):3924–3940. doi:10.1172/JCI127961
39. Grunblatt E, Wu N, Zhang H, et al. MYCN drives chemoresistance in small cell lung cancer while USP7 inhibition can restore chemosensitivity. Genes Dev. 2020;34(17–18):1210–1226. doi:10.1101/gad.340133.120
40. Liu K, Wang S, Liu Y, et al. Overexpression of MYCN promotes proliferation of non-small cell lung cancer. Tumour Biol. 2016;37(9):12855–12866. doi:10.1007/s13277-016-5236-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.