Back to Journals » OncoTargets and Therapy » Volume 13
CircNOL10 Acts as a Sponge of miR-135a/b-5p in Suppressing Colorectal Cancer Progression via Regulating KLF9
Authors Zhang Y, Zhang Z, Yi Y, Wang Y, Fu J
Received 11 December 2019
Accepted for publication 1 May 2020
Published 8 June 2020 Volume 2020:13 Pages 5165—5176
DOI https://doi.org/10.2147/OTT.S242001
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Yuhao Zhang,* Zhijin Zhang,* Yi Yi, Yuexia Wang, Jun Fu
Department of General Surgery, Shanghai No.8 People’s Hospital, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jun Fu
Department of General Surgery, Shanghai No.8 People’s Hospital, No. 8 Caobao Road, Xuhui District, Shanghai 200232, People’s Republic of China
Tel +86-21-34284588
Email [email protected]
Background: Circular RNAs (circRNAs) have been documented as key regulators during progression of malignant human cancer, including colorectal cancer (CRC). However, the underlying molecular mechanisms of circNOL10 in CRC remain unclear.
Methods: The real-time quantitative polymerase chain reaction was used to quantify the expression of circNOL10, miR-135a-5p, miR-135b-5p, and Krüppel-like factor 9 (KLF9). Kaplan–Meier curve was employed to assess the relationship between survival time of CRC patients and expression level of circNOL10. Cell ability of proliferation was measured by Cell Counting Kit8 and colony formation assays. Cell-cycle analysis was performed using flow cytometry assay. In addition, migration and invasion of CRC cell were examined with transwell analysis. The protein expression level was measured with Western blot assay. The interaction relationship of different molecules was analyzed by bioinformatics database and confirmed by dual-luciferase reporter, RNA immunoprecipitation, and RNA pulldown assay. The functional role of circNOL10 in vivo was determined by xenograft experiment.
Results: CircNOL10 was decreased in CRC tissues and cells and was associated with poor outcomes. Gain-of-functional experiment revealed that overexpression of circNOL10 constrained proliferation, cell-cycle progression, migration, and invasion of CRC cells, which was abolished by overexpression of miR-135a-5p or miR-135b-5p. Additionally, miR-135a-5p and miR-135b-5p, targets of circNOL10, regulated KLF9 expression in a negative feedback. Consistently, the results of xenograft experiment suggested that overexpression of circNOL10 inhibited tumor growth in vivo.
Conclusion: In summary, our results showed that circNOL10 impeded CRC development by mediating proliferation, cell cycle, migration, and invasion by sponging miR-135a-5p and miR-135b-5p, which provided new understanding for CRC treatment.
Keywords: circular RNA, circNOL10, miR-135a-5p, miR-135b-5p, KLF9, CRC
Introduction
Colorectal cancer (CRC) ranks third for cancer-related mortality throughout the world, resulting in approximately 880,000 deaths in 2018.1 The poor outcomes of CRC patients partially attributed to lack of diagnostic markers and therapeutic targets, especially in the late stage.2,3 Understanding the molecular mechanisms of metastasis and recurrence in CRC is significant for the improvement prognosis of CRC patients.4
Circular RNAs (circRNAs), covalently closed loop structure without 5ʹcaps and 3ʹ tails, were a group of noncoding RNAs.5 Recently, a large number of evidence had shown that circRNAs acted as modulators in various cancers.6,7 Research by Yang et al claimed that circ-Amotl1 could enhance tumorigenesis progression of breast cancer by binding to c-myc.8 A recent study has shown that 13,198 circRNAs were identified as distinct between CRC and neighboring no-tumor tissues,9 revealing that circRNAs have great potential for CRC diagnosis. CircNOL10 (has_circ_0000977) is derived from the NOL10 gene and located on chr2:10,784,445–10,808,849. CircNOL10 has been investigated to be connected with the process of lung cancer.10 Nevertheless, the mechanism of circNOL10 was not so clearly documented in CRC.
MiR-135 has two subtypes: 135A and 135B, and the sequence of miR-135a-5p and miR-135b-5p was highly conserved. The increased miR-135a/b level was observed in colorectal adenomas and carcinomas.11 In addition, hsa-miR-135a/b could play an important role in the drug-resistance in lung cancer cells.12 Further study revealed that upregulation of miR-135a obviously enhanced proliferation and invasion, while downregulation of miR-135a decreased CRC cell proliferative and invasive capability.13 Therefore, miR-135 might act as optimized strategies for treatment CRC.
Krüppel-like factor 9 (KLF9) was a member of SP/KLF family of DNA-binding transcriptional regulators featured by a highly homologous C-terminal three C2H2-type zinc finger DNA-binding domains.14,15 An emerging body of evidence indicated that Krüppel-like factors (KLFs) were associated with various types of cancers. For example, KLF6 protein was decreased and obviously associated with tumor size.16 Dang et al found that KLF5 mRNA was decreased in breast carcinoma than in matched normal tissues.17
Currently, the study was designed to assess the abundance of circNOL10 in CRC tissues and paired no-tumor tissues. Additionally, we also explored the role and regulatory mechanism of circRNA circNOL10 in CRC in relation to the miR-135a/b-5p/KLF9 axis, while may help us to understand the mechanism of CRC.
Materials and Methods
Human Specimen Tissues
In total 55 paired samples of CRC tissues (32 patients with lymph node metastases and 22 patients without lymph node metastases) and paired non-tumorous tissues were harvested from CRC patients, who had undergone surgical procedure at Shanghai No.8 People’s Hospital. The written informed consents were acquired from all the patients in this study, and that this was conducted in accordance with the Declaration of Helsinki. Patients with CRC were assigned to different groups as Stage I+II or III+IV according to different clinicopathological stages. Moreover, Kaplan–Meier survival curves of CRC patients with high (n=28) or low (n=27) circNOL10 expression were drew with median value of circNOL10 expression as the cutoff. All tissues were frozen in snap-frozen and then transferred to a −80°C until the tissue was processed. The research was authorized by the Ethics Committee of Shanghai No.8 People’s Hospital.
Cell Culture
CRC cells (SW620, SW480, LOVO, and HCT116) were acquired from the American Type Culture Collection (Rockville, MD, USA). Human colonic epithelial cells (NCM460) were gained from Section of Digestive Disease and Nutrition of University of Illinois (Chicago, IL, USA). Those cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Carlsbad, CA, USA) contained 10% fetal bovine serum (FBS; Biochrom KG, Berlin, Germany) and penicillin (50 U/mL; Gibco)-streptomycin (50 mg/mL; Gibco) in a humidified atmosphere contained 5% CO2 at 37°C.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated from cells and tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. For circRNA/mRNA, complementary DNA (cDNA) was synthesized using reverse transcription kit (Takara, Dalian, China), for miRNA, RNA was reversed to cDNA using microRNA Reverse Transcription Kit (Qiagen, Hilden, Germany). The RT-qPCR was performed with the ABI Vii7 system (Applied Biosystems, Foster City, CA, USA), and SYBR Green (Qiagen) was used to dye DNA. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and endogenous small nuclear RNA U6 were selected as housekeeping genes. The transcript levels of circRNA, mRNA, and miRNA were evaluated based on the 2−ΔΔCt method. To analyze resistance of circNOL10 and linear NOL10 mRNA to RNase R, total RNA (10 μg) was incubated with 40 U RNase R (Epicentre Technologies, Madison, WI, USA) at 37°C for 2 h and then used for RT-qPCR. The primers of miR-135a-5p and miR-135b-5p were gained from Qiagen (MS00008624/MS00011130 and MS00003472/MS00001575).
The partial primers were listed:
circNOL10 (F, 5ʹ-CCCAACTCAGGCATGCTTCT-3ʹ; R, 5ʹ-CCCTCTATGGTTCCTGTGGC-3ʹ);
NOL10 (F, 5ʹ-TTTGTTGGTGCAAGTTCTGAAGT-3ʹ; R, 5ʹ-TCCGCAGCATCAGTTTGTAGA-3ʹ);
KLF9 (F, 5ʹ-GCCGCCTACATGGACTTCG-3ʹ; R, 5ʹ-GGATGGGTCGGTACTTGTTCA-3ʹ);
GAPDH (F, 5ʹ-TCCCATCACCATCTTCCAGG-3ʹ; R, 5ʹ-GATGACCCTTTTGGCTCCC-3ʹ);
U6 (Forward, 5ʹ-AACGAGACGACGACAGAC-3ʹ; Reverse, 5ʹ-GCAAATTCGTGAAGCGTTCCATA-3ʹ).
Transfection Assay
For circNOL10 over-expression, human circNOL10 cDNA was amplified and inserted into pCD5-ciR vector (oe-circNOL10; Geneseed Biotech, Guangzhou, China), with empty vector as control. For overexpression of miRNAs, miR-135a-5p or miR-135b-5p mimic was designed and provided from GenePharma (Shanghai, China). CRC cells were sowed in the 6-well plates and cultured at 37°C. When cell confluence reached 60–70%, transfection assay was conducted by transfecting 100 pmol/L of oligonucleotide or 50 nmol/L of vector (lentivirus packaging) into CRC cells using lipofectamine 2000 (Invitrogen) in the light of the producer’s direction. After 48 h of transfection, the cells were collected for the following experiments.
Cell Counting Kit8 (CCK8) Assay
We recorded the proliferation capability of CRC cells every 24 h for 3 times after transfection. In brief, CRC cells at 2×103 were seeded in 96 wells and cultured overnight. After transfection, 10 μL of CCK-8 reagent (Beyotime, Shanghai, China) was added to the cells and then cultured for another 2 h at 37°C. Eventually, the cell viability at 24 h, 48 h, and 72h was measured by measuring optical density at wavelength of 450 nm each well on microplate reader (Applied Biosystems)..
Colony Formation Assay
Transfected CRC cells or control cells were digested by trypsin and diluted into the density of 3000 cells/mL. Two hundred microliters of cell suspension was added into 12-well plate and then cultured. Ten days later, the colonies were fixed with 4% paraformaldehyde and then stained with 0.5% crystal violet solution for 20 min. Colonies containing at least 50 cells were marked and counted for colony formation assay.
Cell-Cycle Analysis
After transfection 48 h, CRC cells were collected and rinsed with 0.5 mL of phosphate buffer saline. Subsequently, 1×106/mL of cells were fixed with 70% ice-cold ethanol overnight at 4°C. Cells were treated with 5 μg/mL RNase R and then stained with 0.25 mg/mL of propidium iodide (PI; Thermo Fisher Scientific, Waltham, MA, USA) and. DNA content was measured on the Moflo XDP (Beckman Coulter, Brea, CA, USA).
Transwell Assay
For the migration assay, homogeneous single CRC cell suspension was added into the upper chamber of 24-well transwell chamber at the density of 1×105 cells per well. The complete medium was added to the bottom chamber (FBS concentration: 10%) to induce cell migration. After incubation at 37°C for 24 h, cells that remained on the upper surface of the membrane were lightly erased, and cells that adhered to the basal side of the membrane were fixed with 95% ethanol and stained with 0.1% crystal violet. Stained cells were observed and counted in random fields per well (100 x) under an inverted microscope (Olympus, Tokyo, Japan). In addition, 24-well transwell chamber was coated with Matrigel (BD Biosciences) for invasion assay.
Western Blot Assay
Proteins from tissue or cells of CC were isolated using lysis buffer (Thermo Fisher Scientific) in accordance with the operation manual. After mensuration for protein concentration, 40 μg of protein was isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein was electroblotted onto polyvinylidene fluoride membranes (GE Healthcare, Piscataway, NJ, USA) and membranes were blocked with 3% Albumin Bovine. Subsequently, membranes were incubated with antibodies anti-cyclinD1 (ab40754; 1:1000 dilution; Abcam, Cambridge, MA, USA), anti-c-myc (ab32072; 1:1000 dilution; Abcam), anti-matrix metalloprotein 9 (MMP9; ab38898; 1:1000 dilution; Abcam), anti-E-cadherin (ab15148; 1:1000 dilution; Abcam), anti-KLF9 (ab227920; 1:1000 dilution; Abcam), and anti-GAPDH (ab181602; 1:1000 dilution; Abcam) overnight at 4°C. The membranes were washed with Tris‑buffered saline with Tween (TBST) three times and then incubated with Goat polyclonal Secondary Antibody to Rabbit IgG-H&L (ab150077; 1:3000 dilution; Abcam) for 1 h at room temperature. ECL Western Blotting Detection Kit (Solarbio, Beijing, China) was used to visualize protein bands and the relative density of the immunoreactive bands was measured by Image J software (National Institutes of Health, Bethesda, MD, USA) and normalized to GAPDH.
RNA Immunoprecipitation (RIP)
RIP experiments were performed using the Magna RIP Kit (Millipore, Bedford, MA, USA). SW620 and SW480 cells were collected and re-suspended in 100 μL of RIP lysis Buffer contained with protease and RNase inhibitors. The cell lysates were related to magnetic beads pre-coated with Ago2 (Millipore) or rabbit IgG (Millipore) at 4°C overnight. After washing off unbound material, the immunoprecipitated RNA was purified and extracted by using the proteinase K buffer and Trizol reagent, respectively. The abundance of circNOL10 was detected using RT-qPCR assay.
Dual-Luciferase Reporter Assay
We predicted the miR-135a/b-5p binding sites of circNOL10 or 3ʹuntranslated region (UTR) of KLF9 using the bioinformatics database starBase (http://starbase.sysu.edu.cn/). The sequences contained the supposed binding sites of miR-135a/b-5p were designed from circNOL10 or 3ʹUTR of KLF9 and inserted into pGL3 vectors (Promega, Madison, WI, USA), named as circNOL10-WT, KLF9 3ʹUTR-WT. Furthermore, the matched interacted sequences were mutated for analysis binding specificity. SW620 and SW480 cells were co-transfected with 0.4 μg of indicated luciferase reporter vectors and 20 pmol of miR-135a/b-5p mimic or NC mimic. After 48 h, the cells were collected for luciferase activity assay using luciferase assay kit (Invitrogen).
RNA Pulldown
For the RNA pulldown assay, SW620 and SW480 cells were infected with 3ʹ-biotinylated miR-135a-5p mimic or miR-135b-5p (Millipore). After transfection 48 h, approximately 1 × 107 cells were lysed and incubated with streptavidin-coupled beads at room temperature for 2 h to form biotin-miRNA-lncRNA complexes, while bio-NC (Millipore) was used as a negative control. After that, RNA was extracted and purified with Trizol reagent and proteinase K, respectively. The abundance of circNOL10 was measured with RT-qPCR assay.
In vivo Experiment
Four- to six-week-old BALB/c nude mice were bought from Shanghai Experimental Animal Center (Shanghai, China). The animal experiment was permitted by Shanghai No.8 People’s Hospital. Animal experiments complied with the ARRIVE guidelines and followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Twenty male BALB/c nude mice were divided into four groups (n=5) and subcutaneously injected CRC cells (SW620 and SW480 cells) stably transfected with vector or oe-circNOL10 by lentiviral vectors. The tumor size was monitored by examining the volume (V = 1/2× ab2 method (length (a) and width (b) length of the tumor)) with calipers weekly. Twenty-eight days after injection, mice were sacrificed and the tumors were collected for the next analysis.
Statistical Analysis
Statistical analysis was conducted using the SPSS 21.0 software (IBM, Somers, NY, USA) based on Student’s t-test or one-way analysis of variance, and P-value less than 0.05 meant significant difference. All data were exhibited as mean ± standard deviation. Pearson’s correlation analysis was performed to test the correlation between the expression levels of miR-135a/b-5p and circNOL10 or KLF9.
Results
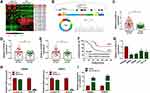
CircNOL10 Was Overexpressed in CRC Tissues and Cells and Was Associated with Poor Prognosis
The analysis results of circRNA microarray (deposited at the GEO accession: GSE126094; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126094) showed that circNOL10 was dramatically decreased in CRC tissues than that in adjacent normal tissues (Figure 1A). Furthermore, circNOL10 was comprised of exons of its parental gene NOL10, as presented in Figure 1B. Consistently, we also confirmed that circNOL10 was downregulated in CRC tissues when compared with adjacent normal tissues (Figure 1C). In addition, the expression levels of circNOL10 were associated with clinicopathological stages and lymphatic metastasis of CRC patients (Figure 1D and E). Importantly, CRC patients with high circNOL10 expression had longer survival time than those patients with low circNOL10 expression (Figure 1F). We also noticed that circNOL10 was decreased in CRC cells (SW620, SW480, LOVO, and HCT116) compared with NCM460 cells (Figure 1G). Moreover, circNOL10 was resistant to RNase R, revealing that circNOL1 was circular (Figure 1H). It was shown that the level of circNOL10 was notably increased in SW620 and SW480 cells transfected with oe-circNOL10 compared with the vector group (Figure 1I). All data indicated that circNOL10 played an important role in CRC progression.
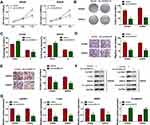
Overexpression of circNOL10 Ameliorated the Transformed Characteristics of CRC Cells
The results of CCK8 assay and colony formation assay indicated that proliferation ability was declined in SW620 and SW480 cells after transfection with oe-circNOL10 (Figure 2A and B). In addition, overexpression of circNOL10 prolonged G0/G1, but shortened S phase in SW620 and SW480 cells (Figure 2C). Transwell assay revealed that circNOL10 overexpression restrained cell migration and invasion (Figure 2D and E). We also found that cyclinD1, c-myc, and MMP9 were decreased but E-cadherin was increased in SW620 and SW480 cells transfected with oe-circNOL10 than cells transfected with vector (Figure 2F and G). We could conclude that overexpression of circNOL10 served as a carcinoma inhibitor in CRC.
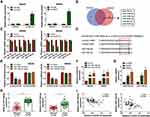
miR-135a-5p and miR-135b-5p Were Overexpressed in CRC Tissues and Cells and Were Targets of circNOL10
By performing RIP assay, circNOL10 was enriched in Ago2 precipitates than that in IgG group (Figure 3A). Interestingly, we found that 6 miRNAs were targets of circNOL10 by prediction with starBase (http://starbase.sysu.edu.cn/) and circBank (http://www.circbank.cn/), including miR-135a-5p and miR-135b-5p (Figure 3B). Moreover, upregulation of circNOL10 inhibited miR-135a-5p and miR-135b-5p expression in SW620 and SW480 cells (Figure 3C). Occasionally, circNOL10 contained putative binding site on miR-135b-5p binding site by performing online bioinformatics starBase analysis (Figure 3D). The data of dual-luciferase reporter assay indicated that all miR-135a-5p or miR-135b-5p mimic could decrease luciferase activity of circNOL10-WT, whereas circNOL10-MUT did not show notable change (Figure 3E). After pulldown, circNOL10 expression of the bio-miR-135a-5p and bio-miR-135b-5p groups was remarkably higher than that of the bio-NC group (Figure 3F). We also observed that miR-135a-5p and miR-135b-5p were overexpressed in CRC tissues and cells than matched controls (Figure 3G and H). Furthermore, miR-135a-5p or miR-135b-5p was negatively correlated with circNOL10 expression in CRC tissues (Figure 3I). Collectively, miR-135a-5p and miR-135b-5p was targets of circNOL10 in CRC.
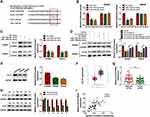
Upregulation with miR-135a-5p or miR-135b-5p Reversed Effects on Proliferation, Cell Cycle, Migration, and Invasion of CRC Cells Caused by Overexpression of circNOL10
To analyze the function of miR-135a-5p or miR-135b-5p in CRC, SW620 and SW480 cells were transfected with NC mimic, miR-135a-5p or miR-135b-5p mimic for further analysis. As shown in Figure 4A, miR-135a-5p or miR-135b-5p was increased in SW620 and SW480 cells transfected with miR-135a-5p or miR-135b-5p mimic with respect to controls, respectively. CCK8 assay and colony formation assay showed that overexpression of circNOL10 impeded proliferation ability of SW620 and SW480 cells, which was overturned after transfection with miR-135a-5p or miR-135b-5p mimic (Figure 4B and C). The flow cytometry assay revealed that overexpression of circNOL10 obviously increased the proportion of cells in G0/G1 phase but decreased the proportion of cells in S phase when compared with controls, whereas upregulation of miR-135a-5p or miR-135b-5p inverted these effects in SW620 and SW480 cells (Figure 4D). Co-transfection of miR-135a-5p or miR-135b-5p mimic and oe-circNOL10 could counteract the oe-circNOL10-induced suppression effects on migration and invasion (Figure 4F). In addition, cyclinD1, c-myc, and MMP9 were declined but E-cadherin was upregulated in SW620 and SW480 cells transfected with oe-circNOL10, which was inverted by overexpression of miR-135a-5p or miR-135b-5p (Figure 4G and H). In summary, overexpression of circNOL10 regulated proliferation, cell cycle, migration, and invasion of CRC cells by targeting miR-135a-5p and miR-135b-5p.
KLF9 Was Bound and Negatively Regulated by miR-135a-5p or miR-135b-5p
As presented in Figure 5A, 3ʹUTR of KLF9 possessed binding sites for miR-135a-5p or miR-135b-5p. Besides, the results of dual-luciferase reporter assay indicated that luciferase activity was declined in SW620 and SW480 cells co-transfected with miR-135a-5p mimic, or miR-135b-5p mimic and KLF9 3ʹUTR-WT than control, whereas co-transfection with miR-135a-5p or miR-135b-5p mimic and KLF9 3ʹUTR-MUT had no effect on the luciferase activities than control (Figure 5B). Moreover, the results of Western blot assay suggested that miR-135a-5p or miR-135b-5p negatively regulated the protein expression of KLF9 in SW620 and SW480 cells (Figure 5C). The inhibition effects on KLF9 expression caused by the circNOL10 overexpression were abolished in SW620 and SW480 cells by upregulation of miR-135a-5p or miR-135b-5p (Figure 5D). We noticed that protein expression of KLF9 was repressed in SW620 and SW480 cells than NCM460 cells (Figure 5E). Importantly, KLF9 expression was declined in CRC tissues (T) compared with normal samples (N) from the TCGA COAD dataset (http://gepia.cancer-pku.cn/detail.php) (Figure 5F). As displayed in Figure 5G and H, KLF9 was decreased in CRC tissues compared with paired normal samples, no matter mRNA or protein. Eventually, correlation assay results revealed that a positive correlation between circNOL10 and KLF9 expression existed in CRC samples (Figure 5I). On the whole, circNOL10 regulated KLF9 expression by targeting miR-135a-5p and miR-135b-5p in CRC.
CircNOL10 Overexpression Decreased Tumor Growth in vivo
To assess the anti-cancer function of circNOL10 in vivo, SW620 and SW480 cells were stably transfected oe-circNOL10 and vector. After CRC cell injection for 4 weeks, tumor volume and weight were remarkably repressed in oe-circNOL10 groups than that in vector groups (Figure 6A and B). As we expect, circNOL10 was overexpressed but miR-135a-5p and miR-135b-5p were declined in oe-circNOL10 groups than that in vector groups (Figure 6C and D). Besides, the results of Western blot analysis revealed that upregulation of circNOL10 impeded cyclinD1, c-myc, and MMP9 expression, whereas KLF9 and E-cadherin were enhanced by circNOL10 overexpression (Figure 6E). Conclusively, circNOL10 exerted function as a tumor inhibitor in CRC.
Discussion
Currently, our results indicated circNOL10 was decreased in CRC tissues and cells than controls. Moreover, circNOL10 was implicated in worse outcomes of CRC patients. Functional experiments revealed that circNOL10 acted as a molecule sponge for miR-135a/b-5p to regulating KLF9 expression, further affected cell proliferation, cell cycle progression, migration, and invasion of CRC cell. CircRNAs related to malignant tumor have been studied in recent years, meanwhile, competing endogenous RNAs (ceRNAs) hypothesis had been acknowledged to function important roles in pathological conditions, especially in cancer occurrence.18,19 Previous studies have also shown close associations between circRNAs and CRC. For instance, Yuan et al indicated that circ_0026344 as a miRNA sponge for miR-21 and miR-31 to impede growth and invasion of CRC cell in vitro.20 What’s more, Nan et al pointed out that circNOL10 inhibiting lung cancer progression by affecting mitochondrial function, accompanying inhibition of proliferation and cell cycle progression as well as enhancement of apoptosis,10 suggesting the anti-tumor potential of circNOL10. Not surprisingly, circNOL10 was confirmed to repress CRC development by acting as a tumor inhibitor in the current study.
Additionally, based on previous conclusions,21 our results also confirmed that circNOL10 was more resistant to RNase R and exhibited higher steadiness than linear NOL10 mRNA. Analysis of functional experiments indicated that overexpression of circNOL10 distinctly constrained CRC cell proliferation and invasion while led to cell cycle arrest. Collectively, circNOL10 exerted its anti-tumor function in CRC by mediation of proliferation, invasion, and cell cycle.
Consistent with previous conclusions,11,13 miR-135a-5p and miR-135b-5p were increased in CRC tissues and cells than that in controls. Additionally, miR-135 was reported to be associated with drug sensitivity,22 bone regeneration,23 and cell differentiation.24 Notably, similar tumor enhancement impacts by miR-135 were found in esophageal squamous cell carcinoma.25 Coincidentally, we predicted the function targets of circNOL10 by bioinformatics databases, and the 6 miRNAs were presented from the overlap, including miR-135a-5p and miR-135b-5p. Importantly, we found that upregulation of circNOL10 resulted in low expression of miR-135a-5p and miR-135b-5p. In addition, RIP and RNA pulldown assays indicated that miR-135a-5p and miR-135b-5p were predicted as direct targets of circNOL10. Notably, the adverse function of miR-135 was confirmed by Jiang et al in breast cancer,26 which may be attributed to different environments of cancer and cell-type-specific.27 More importantly, upregulation of miR-135a-5p or miR-135b-5p inverted the inhibitory effects in CRC development caused by overexpression of circNOL10, suggesting that circNOL10 miR-135a/b-5p axis was associated with CRC progress.
As we all know, miRNAs could regulate target gene by translational inhibition and mRNA destabilization in the posttranscriptional level.28 Ren et al revealed that miR-135a was a potential cancerogenic miRNA in malignant melanoma by targeting FOXO1 expression.29 To probe the downstream target of miR-135a/b-5p in CRC, bioinformatics analysis was performed and KLF9 was predicted as a function target of miR-135a/b-5p. In addition, we observed that there was a negative regulatory relationship between miR-135a/b-5p and KLF9. Mechanistically, Li et al confirmed that KLF9 dramatically suppressed gastric cancer cell invasion and metastasis by repressing MMP28 transcription.30 Kang et al asserted that KLF9 was reduced in CRC tumor tissues than matched normal mucosa tissues.31 Not surprisingly, KLF9 was declined in CRC tissues when compared with paired no-tumor tissues. Rescue assays further verified that circNOL10 exerted biological functions by sponging miR-135a/b-5p in CRC. Regrettably, our experiment did not conduct a comprehensive study on the biological effects of KLF9 on CRC. We will explore the role of KLF9 in CRC in our further study. Another important thing worth mentioning was that investigating the mechanism of miR-135 in various types of cancers was necessary.
Synthetically, the present findings suggest that circNOL10 level was downregulated in CRC tissues and cells than controls, as well as elevated circNOL10 repressed development of CRC. It was the first report to reveal circNOL10/miR-135a/b-5p/KLF9 regulatory signaling in CRC progression.
Conclusion
In this study, circNOL10 was downregulated in human CRC tissues and cells than matched control group. In addition, upregulation of circNOL10 can effectively repress proliferation, cell cycle progression, migration, and invasion of CRC cells. Importantly, our study confirmed the complicated regulation that circNOL10 regulated KLF9 expression by sponging miR-135a-5p and miR-135b-5p in CRC, which may imply that circNOL10 can serve as a therapeutic target for CRC patients.
Abbreviations
CRC, colorectal cancer; KLF9, Krüppel-like factor 9.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Yao H, Zhang Z. Standardized diagnosis and treatment of colorectal liver metastasis. Zhonghua Wei Chang Wai Ke Za Zhi= Chin J Gastrointest Surg. 2017;20(7):753–757.
3. Zhang P, Ji D-B, Han H-B, et al. Downregulation of miR-193a-5p correlates with lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol. 2014;20(34):12241. doi:10.3748/wjg.v20.i34.12241
4. Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(04):191–197. doi:10.1055/s-0029-1242458
5. Chen -L-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205. doi:10.1038/nrm.2015.32
6. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98. doi:10.1186/s13045-018-0643-z
7. Zhu L-P, He Y-J, Hou J-C, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37(5). doi:10.1042/BSR20170750.
8. Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609. doi:10.1038/cdd.2017.86
9. Tian J, Xi X, Wang J, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res. 2019;11:5413. doi:10.2147/CMAR.S199436
10. Nan A, Chen L, Zhang N, et al. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1‐mediated transcriptional regulation of the humanin polypeptide family. Adv Sci. 2019;6(2):1800654. doi:10.1002/advs.201800654
11. Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68(14):5795–5802. doi:10.1158/0008-5472.CAN-08-0951
12. Zhou L, Qiu T, Xu J, et al. miR-135a/b modulate cisplatin resistance of human lung cancer cell line by targeting MCL1. Pathol Oncol Res. 2013;19(4):677–683. doi:10.1007/s12253-013-9630-4
13. Zhou W, Li X, Liu F, et al. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin. 2012;44(10):838–846. doi:10.1093/abbs/gms071
14. McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381. doi:10.1152/physrev.00058.2009
15. Dang DT, Pevsner J, Yang VW. The biology of the mammalian krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32(11–12):1103–1121. doi:10.1016/S1357-2725(00)00059-5
16. Cho Y-G, Choi B-J, Song J-W, et al. Aberrant expression of Krüppel-like factor 6 protein in colorectal cancers. World J Gastroenterol. 2006;12(14):2250. doi:10.3748/wjg.v12.i14.2250
17. Dang DT, Bachman KE, Mahatan CS, et al. Decreased expression of the gut‐enriched Krüppel‐like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett. 2000;476(3):203–207. doi:10.1016/S0014-5793(00)01727-0
18. Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
19. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
20. Yuan Y, Liu W, Zhang Y, et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503(2):870–875. doi:10.1016/j.bbrc.2018.06.089
21. Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
22. Golubovskaya VM, Sumbler B, Ho B, et al. MiR-138 and MiR-135 directly target focal adhesion kinase, inhibit cell invasion, and increase sensitivity to chemotherapy in cancer cells. Anti-Cancer Agents Med Chem. 2014;14:18–28.
23. Xie Q, Wang Z, Zhou H, et al. The role of miR-135-modified adipose-derived mesenchymal stem cells in bone regeneration. Biomaterials. 2016;75:279–294. doi:10.1016/j.biomaterials.2015.10.042
24. Honardoost M, Soleimani M, Arefian E, et al. Expression change of miR-214 and miR-135 during muscle differentiation. Cell J. 2015;17:461.
25. Zhang Y, Ren S, Yuan F, et al. miR-135 promotes proliferation and stemness of oesophageal squamous cell carcinoma by targeting RERG. Artif Cells Nanomed Biotechnol. 2018;46(sup2):1210–1219. doi:10.1080/21691401.2018.1483379
26. Jiang D, Zhou B, Xiong Y, et al. miR-135 regulated breast cancer proliferation and epithelial-mesenchymal transition acts by the Wnt/β-catenin signaling pathway. Int J Mol Med. 2019;43(4):1623–1634. doi:10.3892/ijmm.2019.4081
27. Sood P, Krek A, Zavolan M, et al. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci. 2006;103:2746–2751. doi:10.1073/pnas.0511045103
28. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi:10.1016/S0092-8674(04)00045-5
29. Ren J-W, Li Z-J, Tu C. MiR-135 post-transcriptionally regulates FOXO1 expression and promotes cell proliferation in human malignant melanoma cells. Int J Clin Exp Pathol. 2015;8:6356.
30. Li Y, Sun Q, Jiang M, et al. KLF9 suppresses gastric cancer cell invasion and metastasis through transcriptional inhibition of MMP28. FASEB J. 2019;
31. Kang L, Lü B, Xu J, et al. Downregulation of Krüppel‐like factor 9 in human colorectal cancer. Pathol Int. 2008;58(6):334–338. doi:10.1111/j.1440-1827.2008.02233.x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.