Back to Journals » Cancer Management and Research » Volume 12
CircHIPK3 Promotes Gemcitabine (GEM) Resistance in Pancreatic Cancer Cells by Sponging miR-330-5p and Targets RASSF1
Authors Liu Y, Xia L, Dong L, Wang J, Xiao Q, Yu X, Zhu H
Received 20 November 2019
Accepted for publication 6 January 2020
Published 11 February 2020 Volume 2020:12 Pages 921—929
DOI https://doi.org/10.2147/CMAR.S239326
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yunfei Liu, 1 Li Xia, 2 Luo Dong, 1 Jiale Wang, 1 Qiangsheng Xiao, 1 Xiao Yu, 1 Hongwei Zhu 3
1Department of Hepatobiliary and Pancreatic Surgery‖, Third Xiangya Hospital, Central South University, Changsha 410006, People’s Republic of China; 2Department of Endocrinology, Third Xiangya Hospital, Central South University, Changsha, 410006, People’s Republic of China; 3Department of Gastroenterology and Hepatology, Third Xiangya Hospital, Central South University, Changsha, 410006, People’s Republic of China
Correspondence: Xiao Yu
Department of Hepatobiliary and Pancreatic Surgery‖, Third Xiangya Hospital, Central South University, Changsha 410006, People’s Republic of China
Tel/Fax +86 731-88618831
Email [email protected]
Hongwei Zhu
Department of Gastroenterology and Hepatology, Third Xiangya Hospital, Central South University, Changsha, 410006, Republic of China
Tel/Fax +86 731-88618831
Email [email protected]
Background: Pancreatic cancer is one of the most common malignant diseases in the world. Gemcitabine chemotherapy remains the most important clinical treatment. However, research found that pancreatic cancer cells have chemoresistance to gemcitabine and the effect is not satisfactory. Therefore, it is urgent to find an effective early diagnosis and treatment strategy. Circular RNA is one of the most popular prognostic biomarkers in GEM-resistant PC.
Materials and Methods: The present study was designed to evaluate the role of circHIPK3 in PC. The expression of circHIPK3 in PC tissues and cells and its effect on proliferation, migration, invasion, EMT, and apoptosis were investigated in vitro; its effect on tumor xenografts was assessed in vivo. Used bioinformation analysis to predict which miRNAs could potentially interact with circHIPK3, mRNA, and miR-330-5p.
Results: RT-PCR showed that the level of circHIPK3 was increased in PC tumor tissues; moreover, circHIPK3 was also increased in GEM-resistant PC tumors tissues and GEM-resistant PC cells. Sh-circHIPK3 could knockdown circHIPK3 in PANC-1-GEM and SW-1990-GEM and could significantly inhibit cell proliferation, invasion, migration, EMT and enhance cell apoptosis, compare with control group, the tumor xenografts of circHIPK3 knockdown group were significantly smaller. CircHIPK3 served as a sponge for miR-330-5p, and miR-330-5p directly bound to the 3′ UTR of RASSF1 were revealed by dual luciferase assay and RIP in PC cells. CircHIPK3 knockdown of RASSF1 expression could neutralize the cytological function of PC cells by miR-330-5p inhibitor mediated GEM-resistance.
Conclusion: CircHIPK3 promote gemcitabine (GEM) resistance in pancreatic cancer cells by targeting RASSF1 via miR-330-5p and regulates proliferation, invasive, migration, EMT, and apoptosis. Our research revealed that circHIPK3 may be a novel biomarker in GEM-resistant PC and could be used as a prognostic target.
Keywords: pancreatic cancer, circular RNA, gemcitabine resistance, biomarker
Introduction
Pancreatic cancer (PC) is a malignant tumor of the digestive system with high malignancy and poor prognosis. The incidence of PC was 2.5%, and the mortality rate of all cancers was 4.5% in 2018.1,2 As its early symptoms are not obvious, 80% patients with PC were found when they were in advanced stage of the disease. Currently, gemcitabine (GEM) chemotherapy remains the most important clinical treatment.3,4 Because of acquired drug resistance, PC patients have a poor prognosis. Therefore, the molecular mechanism of GEM-resistance needs to be studied further in PC.
In recent years, circular RNAs (circRNAs) are very important in the occurrence and development of tumor and expected to become a biomarker and diagnosis target for tumor treatment.5–7 Predecessors have confirmed that circRNAs are associated with the occurrence, development, and drug resistance of human diseases, such as Alzheimer’s disease, hepatocellular carcinoma, lung cancer, breast cancer, and PC.8–12 Previous studies have shown that circHIPK3 spongs miR-145-5p, miR-124, miR-558, miR-637, miR-193a, miR-338-3p, and miR-637 to regulate oxaliplatin-resistance, proliferation, tube formation, invasion, autophagy, migration, and apoptosis in lung cancer, preeclampsia, gastric cancer, colorectal cancer, chronic myeloid leukemia, bladder cancer, prostate cancer.13–20 However, the role and function of circHIPK3 have not been revealed in PC.
In the present study, we found that the level of circHIPK3 was increased in PC tissues; moreover, circHIPK3 was also increased in GEM-resistant PC tumor tissues and GEM-resistant PC cells. Further investigation demonstrated that circHIPK3 could regulate GEM-resistant PC by targeting the miR-330-5p/RASSF1 axis. Our study revealed that CircHIPK3 may also be a novel biomarker in PC and may be used as a prognostic target.
Materials and Methods
Clinical Human Tissue Samples
The cancer tissues and adjacent tissues of 28 patients with PC were obtained from Third Xiangya Hospital, Central South University during surgical, and then all tissues were rapidly frozen in liquid nitrogen until use. GEM-resistance definition: PC patients have no response by GEM therapy or previous GEM treatment had progression but no more than 6 months.
Cell Lines Culture and Transfection
Human PC cell lines PANC-1 and SW 1990 were purchased from the Cell Collection Committee of the Chinese Academy of Sciences (Shanghai, China) and cultured at 37°C in a 5% CO2 humidified incubator. PANC-1-GEM and SW 1990-GEM were created by previous study.21 According to the manufacture’s protocol, 50 nmoL sh-circHIPK3 or miR-330-5p mimic or negative control (NC) were transfected into PANC-1-GEM and SW 1990-GEM cells using Lipofectamine 2000 (Invitrogen).
RNA Extraction and Real-Time PCR (RT-PCR) Analysis
Total RNA was extracted from samples by TRizol reagent (Invitrogen, USA) with manufacturer’s protocol. Fast-Start Universal SYBR GEMeen Master (Rox) (Roche, Switzerland) was used to carry out real-time quantitative PCR on CFX96Tm Real-Time System (Bio-Rad, USA). CircHIPK3-1-Forward: TATGTTGGTGGATC-CTGTTCGGCA, CircHIPK3-1-Reverse: TGGTGGGTAGACCAAGACTTGTGA; CircHIPK3-2-Forward:CTACAGATCCGACCAGGAGTTC, CircHIPK3-2-Reverse: TGTGAACCAGCCACACTCTCAG; miR-330-5p-forward: TCTCTGGGCCTGT-GTCTTAGGC, miR-330-5p- reverse: GCTATCTCAGGGCTTGTTGCTTCAGTC-CTCCTGGG; RASSF1-Forward: CCCTTGGGTGACCTCTTGTA, RASSF1-Reverse: GCTGTTCTCTGGGCTCATTC; U6-Forward: CTCGCTTCGGCAGCACA, U6-Reverse: AACGCTTCACGAATTTGCGT; GAPDH-Forward: GGGAGCCAA-AAGGGTCAT, GAPDH-Reverse: GAGTCCTTCCACGATACCAA; Each sample repeated 3 parts, the expression of circHIPK3, miR-330-5p, RASSF1 were calculated by 2−∆∆Ct method.
CCK8 Assay
After 36 hrs of transfection, 2×105 cells/well PANC-1-GEM and SW 1990-GEM cells were seeded into 96-well plates with 6 replicate wells in each group. Twenty-microliter CCK-8 reagent (Sigma-Aldrich, USA) was added into 96-well plates, after incubating for 4 hrs without light. The OD value was measured with a microplate detector at 450 nm (EnSpire 2300, PerkinElmer, USA).
Flow Cytometry
Digest the cells with 0.25% EDTA-free trypsin, collected the culture solution of cells each well and have fallen off cells after digestion in a flow tube. Dyeing FITC and PI according to Annexin V-labelled detection kit (Life technologies, USA) manufacturer’s protocol. After 15 mins of incubation in the dark, the rate of apoptosis was detected by flow cytometer (BD Biosciences).
Dual-Luciferase Reporter Assay
First, construction of miR-330-5p and circHIPK3 wild type (WT) or circHIPK3 mutant-type (MUT) plasmids, respectively, the recombinant plasmid was co-transfected into cells with miR-330-5p and NC. After transfection 48 hrs, cells were collected and the luciferase activities were observed according to dual-luciferase Reporter Assay System (Promega) manufacturer’s protocol.
RNA Immunoprecipitation (RIP)
Collecting 1×107 cells and add equal volume of RIP lysate. According to RIPTm RNA binding protein immunoprecipitation reaction kit manufacturer’s protocol. Then, RNeasy Mini Kit (QIAGEN, GER) was used to purify RNA and the expression of miR-330-5p and circHIPK3 were detected by qRT-PCR.
Wound Healing and Transwell Assays
The monolayer cells were scraped by the tip of 200-μL pipette, and then cultured in the medium with 2% FBS. The wound healing was observed under the Inverted microscope (Nikon, Japan) and photos were taken at 0 hr and 24 hrs.
Transwell, the upper chamber was added with Matrigel in advance, and then 200 μL 1×105/mL transfected cells with 2% FBS was seeded and incubated on the upper chamber, the bottom chamber was added 600 μL medium with 20% FBS. After 48 hrs, remove the cells and Matrigel in the upper chamber and fix the bottom chamber of migration cells with formaldehyde. The number of cells were measured by Inverted microscope (Nikon, Japan)
Nude Mouse Tumorigenicity Assay
0.1 mL of shRNA circHIPK3 or sh-NC GEMoup cells (2×107/mL) were injected into the abdominal cavity of BALB/c nude mice. The tumor was visible after 2 weeks and tumor size was observed weekly, as follows: 0.5×W2×L every week.
Western Blot
The total protein was extracted by RIPA buffer, the quantified of protein samples were measured by BCA protein assay kit. All samples were incubated with the primary antibody (RASSF1, E-cadherin, N-cadherin, vimentin, GAPDH, Proteintech) at 4°C for 12 hrs, then incubated with a secondary antibody at 37°C for 2 hrs. Chemiluminescence system (Bio-Rad, USA) imaging system was used to measure all samples.
Statistical Analysis
SPSS 21.0 software was used for statistical analysis. Mean±SD is used to represent measurement data. Student’s t test was used for statistical analysis. Survival rate was analyzed by Kaplan–Meier curve and Log-rank test, the correlation analysis of circHIPK3, miR-330-5p, and RASSF1 by Pearson. P<0.05 represents statistical significance.
Ethics Statement
The study was approved by the Ethics Committee Third Xiangya Hospital of Central South University, and obtained the written informed consent of all participants. Instructive notions with respect to caring for laboratory animals (which is released by the Ministry of Science and Technology of the People’s Republic of China on September 30th, 2006.) were followed for the welfare of the animals.
Results
CircHIPK3 Was Increased in PC Tissues and GEM-Resistant PC Tissues
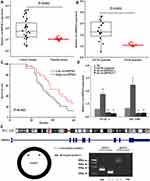
QRT-PCR detected the expression of circHIPK3 in 28 pairs PC tissues and matched normal tissues. Compared with normal tissues, the results showed that the expression of circHIPK3 was up-regulated in PC tissues and GEM-sensitive PC tissues (P<0.01, Figure 1A and B). Further statistical analyses evaluated that the high expression of circHIPK3 displayed poorer survival rate compared with low circHIPK3 in PC patients (P<0.05, Figure 1C).
Compared with parental cell lines, the expression of circHIPK3 was significantly increased in PANC-1-GEM and SW 1990-GEM cells (P<0.05, Figure 1D). Then, we next assessed the exon structure of circHIPK3, which derived from exon 11 of HIPK3 gene. RT-PCR was used to amplify linear and circular RNA HIPK3 based on cDNA and genomic DNA (gDNA) by convergent primers and divergent primers, RT-PCR showed that divergent primers could amplify by in cDNA but not in gDNA (Figure 1E).
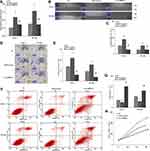
Low Expression CircHIPK3 Suppressed GEM-Resistant PC Cells’ Proliferation, Invasive, Migration, and Enhance Cell Apoptosis
ShRNA circHIPK3 was transfected into PANC-1-GEM and SW 1990-GEM, and sh-circHIPK3 efficiently suppressed circHIPK3 expression in PANC-1-GEM and SW 1990-GEM cells (Figure 1D). Subsequently, compared with the control group, CCK8 assay, wound healing, transwell assays, and flow cytometry revealed that knockdown of circHIPK3 could significantly suppress PANC-1-GEM and SW 1990-GEM cells’ proliferation, invasive, migration, and promoted cell apoptosis (Figure 2A–G). Furthermore, the average volume of tumor xenografts was significantly smaller in the circHIPK3 knockdown group (p<0.05, Figure 2H). These results indicated that down-regulated of circHIPK3 enhance PC cells sensitization to GEM treatment.
CircHIPK3 Interacts with miR-330-5p in PC
Using online database circbank and starBase to predict the miRNAs potentially interacting with circHIPK3. Dual-luciferase reporter assay was performed to identify the miRNAs that binding to circHIPK3 and 4 miRNAs (miR-330-5p, miR-582-3p, miR-142-5p, miR-1179) luciferase activity were significantly decreased compared with the control group (Figure 3A and B), then miR-330-5p was copurified with circHIPK3 by RIP assay (Figure 3C and D). Compared with their parental control, the expressions of miR-330-5p were increased in PANC-1-GEM and SW 1990-GEM cells transfected with sh-circHIPK3 (Figure 3E). Notably, the expressions of miR-330-5p were reduced in PC tumor tissues and GEM-resistant PC tumor tissues (Figure 3F and G). In summary, these results demonstrated that circHIPK3 sponging to miR-330-5p.
CircHIPK3 Promotes Gemcitabine (GEM) Resistance in Pancreatic Cancer Cells by Targeting RASSF1 via miR-330-5p
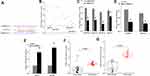
Using online database (targetScan, starbase, picTar, RNA22, and miRanda) to predict mRNA potentially interacting with miR-330-5p, bioinformatics analysis found that miR-330-5p could bind to RASSF1 3ʹ UTR (Figure 4A). The luciferase activity of co-transfected RASSF1 with miR-330-5p-WT was also significantly decreased in PANC-1-GEM cells (Figure 4B and C). miR-330-5p inhibitor could significantly upregulate RASSF1 expression in PANC-1-GEM and SW 1990-GEM cells (Figure 4D). These results confirmed that the target gene of miRNA-429 with RASSF1.
To investigate that whether circHIPK3 exerted its oncogenic role through sponging miR-330-5p. We further knockdown of RASSF1 expression by sh-circHIPK3 (Figure 4E). RT-PCR showed that sh-circHIPK3 treatment could significantly downregulate RASSF1 expression in PANC-1-GEM and SW- 1990-GEM cells, miR-330-5p inhibitor could reverse circHIPK3 knockdown mediated GEM-sensitization indicated by promotion of cell proliferation, invasive, cell apoptosis, and EMT (Figure 4F–L), these results showed that knockdown of circHIPK3 could suppress RASSF1 expression and could neutralize the function of PC cells by miR-330-5p inhibitor mediated GEM-resistance.
In addition, the expressions of RASSF1 were increased in PC tumor tissues and GEM-resistant PC tumor tissues (Figure 4I and J). Correlation analysis showed that circHIPK3 and RASSF1 with miR-330-5p were negatively correlated (Figure 4M). In conclusion, these studies demonstrated that circHIPK3 promotes Gemcitabine (GEM) resistance by targeting RASSF1 via miR-330-5p and regulates proliferation, invasive, migration, EMT, and apoptosis in PC.
Discussion
CircRNA is a type of novel noncoding RNA that is closely associated with carcinogenesis and cancer progression. The function of circRNAs had received much attention.22,23 CircRNAs had been proved to be involved in the development, chemotherapy resistance, proliferation, invasion, and prognosis of PC.24–26 In our study, the level of circHIPK3 was increased in PC tissues, GEM-resistant PC tissues, and GEM-resistant PC cells. PC patients with high expression of circHIPK3 displayed poorer overall survival than those with lower circHIPK3 expression. Research indicates that circHIPK3 was increased in prostate cancer, gastric cancer, and lung cancer. However, circHIPK3 was decreased in gastric cancer tissues. Therefore, circHIPK3 may have some unique features, such as stability, higher expression, and expression specific to tissue developmental stage.27
It is now widely accepted that circRNAs sponging to miRNAs. To explore the mechanism of circHIPK3 in GEM-resistant PC. Bioinformatics analysis, luciferase reporter, and RIP showed that circHIPK3 served as a sponge for miR-330-5p in PC cells. The mutant sequence of miR-330 is miR-330-5p, and several studies reported that miR-330-5p unction as oncogenes or tumor suppressor genes in tumor progression such as breast cancer, lung cancer, prostate cancer, PC, etc.28–31 MiRNAs can regulate tumorogenesis by regulating their target mRNAs, and miR-330-5p directly bound to RASSF1 3′- UTR. Recent evidence indicated that RASSF1 is involved in various biological processes such as apoptotic signaling, microtubule stabilization, and mitotic progression, etc.32,33 Our results showed that RASSF1 expression was significantly upregulated in GEM-resistant PC tissues, GEM-resistant PC cells and knockdown of circHIPK3 were also significantly downregulated RASSF1.
Our study demonstrated that knockdown of circHIPK3 could significantly affect cell proliferation, invasion, migration, apoptosis, and EMT in PC cells. Furthermore, compared with control group, the tumor xenografts of circHIPK3 knockdown group were significantly smaller, and circHIPK3 knockdown-mediated GEM sensitization indicated by promote cell proliferation, migration, invasion, and inhibit cell apoptosis were reversed by miR-330-5p inhibitor. These results showed that downregulation of circHIPK3 promotes PC cell sensitization to GEM treatment and as an oncogenic to targeting the miR-330-5p/RASSF1 axis.
Conclusion
The study showed that circHIPK3 promotes GEM-resistance by acting as a ceRNA to targets RASSF1 by sponging miR-330-5p affected to GEM-resistant PC cell proliferation, migration, invasion, cell apoptosis, and EMT. CircHIPK3 may be a potential target in GEM-resistant PC.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (81873589), and the New Xiangya Talent Project of the Third Xiangya Hospital of Central South University (no.JY20160310).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
3. Oneda O, Zaniboni Z. Are we sure that adjuvant chemotherapy is the best approach for resectable pancreatic cancer? Are we in the era of neoadjuvant treatment? A review of current literature. J Clin Med. 2019;8(11):1922. doi:10.3390/jcm8111922
4. Lin KI, Yang JL, Lin YC, et al. Network meta-analysis of efficacy and safety of chemotherapy and target therapy in the first-line setting of advanced pancreatic cancer. Cancers (Basel). 2019;11(11):1746. doi:10.3390/cancers11111746
5. Ye DX, Wang SS, Huang Y, et al. A 3-circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int. 2019;19:276. doi:10.1186/s12935-019-0995-7
6. Liu Q, Shuai M, Xia Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag Res. 2019;11:8023–8031. doi:10.2147/CMAR.S218967
7. Yu X, Ding H, Yang L, et al. Reduced expression of circRNA hsa_circ_0067582 in human gastric cancer and its potential diagnostic values. J Clin Lab Anal. 2019;e23080.
8. Dube U, Del-Aguila JL, Li Z, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22(11):1903–1912. doi:10.1038/s41593-019-0501-5
9. Wang W, Li Y, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother. 2019;121:109517.
10. Zhao CH, Qu L, Zhang H, et al. Identification of breast cancer-related circRNAs by analysis of microarray and RNA-sequencing data: an observational study. Medicine (Baltimore). 2019;98(46):e18042. doi:10.1097/MD.0000000000018042
11. Cheng X, Qiu J, Wang S, et al. Comprehensive circular RNA profiling identifies CircFAM120A as a new biomarker of hypoxic lung adenocarcinoma. Ann Transl Med. 2019;7(18):442. doi:10.21037/atm
12. Li Q, Geng S, Yuan H, et al. Circular RNA expression profiles in extracellular vesicles from the plasma of patients with pancreatic ductal adenocarcinoma. FEBS Open Bio. 2019;9(12):2052–2062. doi:10.1002/feb4.v9.12
13. Zhang Y, Li C, Liu X, et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288. doi:10.1016/j.ebiom.2019.09.051
14. Chen X, Mao R, Su W. et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy;2019. 1–13. doi:10.1080/15548627.2019.1634945
15. Feng XQ, Nie SM, Huang JX, et al. Circular RNA circHIPK3 serves as a prognostic marker to promote chronic myeloid leukemia progression. Neoplasma. 2019. doi:10.4149/neo_2018_181129N908
16. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi:10.1016/j.bbrc.2018.10.087
17. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/beta-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23(18):7905–7912. doi:10.26355/eurrev_201909_19004
18. Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi:10.15252/embr.201643581
19. Cai C, Zhi Y, Wang K, et al. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi:10.2147/OTT.S196931
20. Chen D, Lu X, Yang F, et al. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. 2019;11:1415–1423. doi:10.2147/CMAR
21. Li D, Qian X, Xu P, et al. Identification of lncRNAs and their functional network associated with chemoresistance in SW1990/GZ pancreatic cancer cells by RNA sequencing. DNA Cell Biol. 2018;37(10):839–849. doi:10.1089/dna.2018.4312
22. Teng F, Xu J, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;112:8–17. doi:10.1016/j.biocel.2019.04.011
23. J H X, Wang Y, Xu D. Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomark. 2019;25(2):193–201. doi:10.3233/CBM-182293
24. Chen Y, Li Z, Zhang M, et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2019;38(1):466. doi:10.1186/s13046-019-1436-0
25. Zhang Q, Wang JY, Zhou SY, et al. Circular RNA expression in pancreatic ductal adenocarcinoma. Oncol Lett. 2019;18(3):2923–2930. doi:10.3892/ol.2019.10624
26. Xu Z, Shen J, Hua S, et al. High-throughput sequencing of circRNAs reveals novel insights into mechanisms of nigericin in pancreatic cancer. BMC Genomics. 2019;20(1):716. doi:10.1186/s12864-019-6032-3
27. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
28. Su BB, Zhou SW, Gan CB, et al. MiR-330-5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. J Surg Res. 2016;203(2):434–440. doi:10.1016/j.jss.2016.03.021
29. Kong R, Liu W, Guo Y, et al. Inhibition of NOB1 by microRNA-330-5p overexpression represses cell growth of non-small cell lung cancer. Oncol Rep. 2017;38(4):2572–2580. doi:10.3892/or.2017.5927
30. Liu DC, Song LL, Liang Q, et al. Long noncoding RNA LEF1-AS1 silencing suppresses the initiation and development of prostate cancer by acting as a molecular sponge of miR-330-5p via LEF1 repression. J Cell Physiol. 2019;234(8):12727–12744. doi:10.1002/jcp.v234.8
31. Gao J, Wang G, Wu J, et al. Skp2 expression is inhibited by arsenic trioxide through the upregulation of miRNA-330-5p in pancreatic cancer cells. Mol Ther Oncolytics. 2019;12:214–223. doi:10.1016/j.omto.2019.01.006
32. Arnette C, Efimova N, Zhu X, et al. Microtubule segment stabilization by RASSF1A is required for proper microtubule dynamics and Golgi integrity. Mol Biol Cell. 2014;25(6):800–810. doi:10.1091/mbc.e13-07-0374
33. Fu L, Zhang S. RASSF1A promotes apoptosis and suppresses the proliferation of ovarian cancer cells[J]. Int J Mol Med. 2014;33(5):1153–1160. doi:10.3892/ijmm.2014.1671
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.