Back to Journals » OncoTargets and Therapy » Volume 14
circHIPK3 (hsa_circ_0000284) Promotes Proliferation, Migration and Invasion of Breast Cancer Cells via miR-326
Authors Qi L, Sun B, Yang B, Lu S
Received 4 January 2021
Accepted for publication 18 March 2021
Published 9 June 2021 Volume 2021:14 Pages 3671—3685
DOI https://doi.org/10.2147/OTT.S299190
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Liqiang Qi,1 Bo Sun,2 Beibei Yang,2 Su Lu2
1Department of Breast Surgical Oncology, Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, People’s Republic of China; 2The 2nd Department of Breast Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China
Correspondence: Liqiang Qi
Department of Breast Surgical Oncology, Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, No. 17, Panjiayuan Nanli, Chaoyang District, Beijing, 100021, People’s Republic of China
Tel +86-10-67781331
Email [email protected]
Purpose: circHIPK3 has carcinogenic or anti-tumor effects on different cancers. However, there is no relevant research showing whether circHIPK3 was involved in breast cancer (BCa). In this research, the aim was to analyze the function and possible molecular mechanism of circHIPK3 in BCa.
Methods: The expression of circHIPK3 in human BCa tissues and cells was detected by real-time quantitative PCR (RT-qPCR). CircInteractome and dual-luciferase assays were performed to detect circRNA–miRNA targeting relationship. Ribonuclease R treatment, RT-qPCR, Western blot and immunohistochemistry were performed to determine the stability, expressions, abundance of target genes. Loss-of-function or gain-of-function experiments were used to analyze the effects of circHIPK3 and miR-326 on BCa in vivo and in vitro. In vitro, MCF7 and BT20 cells were transfected with circHIPK3 or sicircHIPK3 or miR-326 mimic; in vivo, female BALB/c mice were subcutaneously injected with MCF7 cells (transfected with CirchipK3 or miR-326 mimic) to establish xenograft models.
Results: The circular structure of circHIPK3 was abundantly expressed in the cytoplasm and was up-regulated in BCa. Silenced circHIPK3 suppressed malignant phenotype of BCa cells. MiR-326 interacted with circHIPK3 and the two were negatively correlated. Overexpressed circHIPK3 promoted cell viability, proliferation, migration and invasion, but inhibited apoptosis. Moreover, overexpressed circHIPK3 promoted the expressions of EMT-related genes and antiapoptotic genes, but inhibited proapoptotic gene expressions. Overexpressed circHIPK3 promoted tumor growth and Ki-67 levels, inhibited apoptosis in vivo. The above mentioned effects of circHIPK3 were reversed by miR-326 in vitro or in vivo.
Conclusion: circHIPK3 promoted proliferation, migration and invasion of BCa cells through regulating miR-326.
Keywords: breast cancer, circRNA, circHIPK3, miR-326, malignant progress
Introduction
Lung cancer, breast cancer (BCa) and colon cancer are the three most common tumors in women.1 The number of BCa deaths reached 41,760 cases, accounting for 15% of the cancer-related death in 2019, and its 5-year relative survival rate was 90%.1 Surgery is currently the main treatment for BCa, but BCa is metastatic and aggressive, and patients are prone to relapse again.2 In addition, the prognosis of patients is largely difficult to predict, due to the complex pathogenesis of BCa.3,4 Therefore, studying the molecular mechanism of BCa progression and developing novel early diagnostic biomarkers and therapeutic targets are of great significance for an effective treatment of BCa.
The discovery of circRNA changed the traditional cognitive view of non-coding RNA encoding protein in the body.5 Previous studies have noted that the importance of circRNAs in various physiological and pathological processes.6 In practice, Ma et al found that circRNA-0000284 can interact with miR-506 to regulate the proliferation and invasion of cervical cancer cells.7 Zhang et al found that the expression level of circ_101222 is significantly different between preeclampsia women and normal women.8 Currently, a large number of studies have found that circRNA is involved in regulating the molecular characteristics of BCa and the clinical manifestations of BCa patients.9 Zheng et al detected at least 27,000 circRNAs including circHIPK3 and differentially expressed genes from normal tissues and BCa tissues.10 In addition, they pointed out that circHIPK3 was found to directly bind to miR-124 and inhibit miR-124 activity.10 The expression of circIRAK3 is upregulated in metastatic BCa cells and can promote the migration and invasion of BCa cells; circIRAK3 combines with miRNA-3607 to upregulate the expression of forkhead box C1, thereby promoting the metastasis of BCa cells in vivo.11
CircHIPK3 (hsa_circ_0000284) is widely distributed, rich in content, and stable in structure.12,13 A number of studies proved that circHIPK3 has carcinogenic or anti-tumor effects on different cancers such as ovarian cancer, colorectal cancer and liver cancer.14–16 Zhang et al pointed out that circHIPK3 knockdown can improve fibroblast-to-myofibroblast transition and inhibit fibroblast proliferation by regulating miR-338-3p.17 However, whether circHIPK3 was abnormally expressed in BCa was unclear. In this investigation, the aim was to determine the expression, function and possible molecular mechanism of circHIPK3 in BCa.
Materials and Methods
Ethics Statement
BCa tissues and adjacent tissues (ANT) (n = 48) were selected from Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College from Jan. 2020 to Jul. 2020 and were approved by the Ethics Committee of Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (BL201912020). All patients had signed and provided informed consent, and this was conducted in accordance with the Declaration of Helsinki. The animal assays were approved by the Institutional Animal Care and Use Committee of Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, with approval number: BN202001015. All animal experiments were performed in accordance with the guidelines of the China Council on Animal Care and Use.
Cell Culture
Human normal mammary epithelial cell line MCF-10A (CRL-10,317), and BCa cell lines MCF7 (HTB-22), SK-BR-3 (HTB-30), BT549 (HTB-122), BT20 (HTB-19), MDA-MB-231 (HTB-26), and MDA-MB-453 (HTB-131) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (PM150210A, Procell, China) with 10% Fetal Bovine Serum (FBS, 164,210, Procell) at a 37°C, 5% CO2 incubator (SCO6WE-2, SHELLAB, USA). All cell lines were purchased from American Type Culture Collection (ATCC, USA).
Ribonuclease R (RNase R) and Actinomycin D Treatment
For RNase R resistance detection, total RNA of circHIPK3 and HIPK3 (2 μg) were incubated with 3U/μg RNase-R (07250, Epicentre Technologies, USA) at 37°C for 30 minutes (min). For half-life detection, the cell medium was added to 2 mg/mL Actinomycin D (129,935, Millipore, USA) to block transcription. After treatment, the expressions of circHIPK3 and HIPK3 were determined by real-time quantitative PCR (RT-qPCR).
Transfection
Overexpressed circHIPK3 (circHIPK3), small interfering RNAs (siRNAs) targeting circHIPK3 (sicircHIPK3) and corresponding negative control (siNC or NC) recombinant plasmids were obtained from Guangzhou GENESEED company (China). CircHIPK3 junction site: GGTACTACAGGTATGGCCTCA. MiR-326 mimic (M, miR10000756-1-5), and mimic control (MC, miR1N0000002-1-5) was obtained from RIBOBIO Co., Ltd. (Guangzhou, China). The Lipofectamine 3000 (L3000015, Invitrogen, USA) were employed to transfect MCF7 or BT20 cells. Briefly, MCF7 or BT20 cells (1 x 106) were seeded into 6-well plate, and after cells reached 70% confluence, cells transfection was performed. Lipofectamine 3000 reagent was diluted with Opti-MEM medium, and sicircHIPK3, siNC, circHIPK3, NC, miR-326 mimic and mimic control were diluted with Opti-MEM medium. The two dilutions were then mixed in a 1:1 ratio and incubated at room temperature for 5 min. The liposome complex was then added to the cells and incubated at 37°C for 2 days. Finally, the cells were collected for further analysis.
RNA Isolation and RT-qPCR
Cytoplasmic & Nuclear RNA Purification Kit (NGB-21,000, NorgenBiotek, Canada) was used in nuclear and cytoplasmic RNA extraction. Total RNAs from BCa cells or tissues were isolated using RNA Extraction Kit (LS1040, Promega, USA). RT-PCR kit (T2240, Solarbio, China) and miRcute Plus miRNA First-Strand cDNA Kit (KR211-01, TIANGEN, Beijing, China) were employed to generate cDNA. The TB Green Fast qPCR Mix (RR430A, Takara, China) and miRcute Plus miRNA qPCR Kit (SYBR Green) (FP411-02, TIANGEN, Beijing, China) were used to perform RT-qPCR in ABI 7500 real-time PCR system (Applied Biosystems, USA). The expressions of target genes were calculated by the 2−ΔΔCt method,18 and GAPDH or U6 served as control. All the primer sequences are listed in Table 1.
 |
Table 1 All Primers in This Study |
Cell Viability Assay
BCa cells (about 1×104/well) were incubated regularly for 24, 48 or 72 hours (h). Next, cell counting kit-8 (CCK-8) reagents (10 μL) (HY-K0301, MedChemExpress, USA) were added and incubated for 2 h. Thereafter, the OD value at 450 nm was analyzed with an EnVision microplate reader (PerkinElmer, USA).
Colony Formation Assay
BCa cells (1 ×102/well) were inoculated into 6-well plates and incubated regularly for about 2 weeks. Thereafter, the colonies were fixed with 4% paraformaldehyde (16,005, Sigma-Aldrich, USA) for 15 min and stained with 0.1% crystal violet (V5265, Sigma-Aldrich, USA) for 15 min. Finally, the numbers of colonies were analyzed by a Nikon ECLIPSE Ts2 microscope (Japan).
Wound-Healing Assay
BCa cells were transfected with sicircHIPK3, siNC, circHIPK3, NC, miR-326 mimic and mimic control in 100 mm petri dish. After cell collection, the cell density was adjusted to 5×105/mL and seeded into 35 mm petri dish and incubated regularly until approximately 90% confluence. Then, a pipette tip was used to create a straight line wound on cell layers. At 48 h after culture, the cell images were analyzed with a microscope (magnification × 100). Relative migration rate of cells was analyzed by Image J (version 1.48, National Institutes of Health), relative migration rate = (0 h scratch width − 48 h scratch width)/0 h scratch width × 100%.
Transwell Assay
Firstly, the Transwell chamber (3422, Corning, USA) was covered with Matrigel (YZ-356,234, Solarbio, China) to form an artificially restructured basement membrane. Then, the medium containing 10% FBS was added to the 24-well plate. The Transwell chamber was placed in the 24-well plate, and added with 100 μL of BCa cell suspensions (1 × 103 cells/well). After 48 h, the chamber was taken out and the cells on the inner surface of the filter were wiped off. Finally, the invading cells were fixed by formaldehyde (F8775, Sigma-Aldrich, USA), stained with crystal violet, and ultimately analyzed under a microscope (magnification × 250).
Flow Cytometry
Annexin V-FITC Apoptosis Detection Kit (CA1020) was purchased from Solarbio (China). The pre-digested BCa cells (1×105/well) were mixed with 5 µL of Annexin V-FITC for 10 min, followed by mixing with 5 µL of propidium iodide staining solution for 5 min. Afterwards, CytoFLEX flow cytometry (Beckman Coulter, USA) was performed for the detection of cell apoptosis.
Bioinformatics Prediction and Dual-Luciferase Assay
CircInteractome (https://circinteractome.nia.nih.gov/) was used to screen circHIPK3 targeted miRNAs (Supplementary Table). Dual-luciferase assay was constructed for target gene verification. The sequences of circHIPK3 containing wild-type or mutant miR-326 binding sites (circHIPK3-WT or circHIPK3-MUT) were synthesized and sub-cloned into pmirGLO reporter plasmids (E1330, Promega, USA). BCa cells were co-transfected with circHIPK3-WT or circHIPK3-MUT, and with miR-326 inhibitor (miR20000756-1-5, RIBOBIO, China) or inhibitor control (miR2N0000001-1-5, RIBOBIO, China). After transfection for 48 h, dual-luciferase activities were determined by TransGen Biotech luciferase detection kit (FR201-01, China) and a GENios Pro plate reader (Tecan, Switzerland).
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using the EZ-Magna RIP RNA-Binding Protein Immunoprecipitation kit (17–701, Millipore, USA), according to the manufacturer’s instructions. MCF7 and BT20 cells were lysed in complete RIP lysis buffer, and then incubated with RIP buffer containing magnetic beads conjugated with Anti-Ago2 antibody and normal negative control IgG (Millipore) for 6 h at 4°C. circHIPK3 and miR-326 enrichment in the precipitation were detected by RT-qPCR assay.
Western Blot
Total protein extraction kit (BC3711, Solarbio, China) was performed for extraction of total protein from BCa cells or tumor tissues. The concentrations of total proteins were measured with BCA kit (P0011, Beyotime, China), and separated with 10% SDS-PAGE gel. After that, the proteins were transferred onto the PVDF membrane (160–0184, Bio-Rad, USA) and then sealed for 2 h. Thereafter, the membranes were incubated with corresponding primary antibodies at 4°C for overnight, followed by incubation with secondary antibodies at room temperature for 2 h. Subsequently, the bands were visualized with BeyoECL Plus (P0018M, Beyotime) and analyzed with Image J 1.42 software (USA). Antibodies information were listed in Table 2.
 |
Table 2 All Antibodies Information and Sources in This Study |
Tumorigenesis Assay
Thirty-two female BALB/c mice (4–6 weeks old, N000020) were obtained from Nanjing Information Technology Center of Gempharmatech (China). All the mice were raised in SPF-level animal laboratory with 22–25°C and 55±5% humidity, and provided with free access to food and water.
NC + MC, NC + M, circHIPK3+ MC, circHIPK3 + M groups were transfected into MCF7 cells and prepared into cell suspension (2×105/mL) for use. Female BALB/c mice were randomly divided into four groups (n = 3), and the same amount of different groups of MCF7 cells were subcutaneously injected into the mice to establish xenograft models. Tumor formation was frequently observed. Then, about four weeks later, the mice were euthanized (sodium pentobarbital, 150 mg/kg, intraperitoneal, P3761, Sigma-Aldrich, USA). Afterwards, the tumor was collected, and the weight was calculated. Images were captured, and the tumor tissues were quickly frozen or fixed for later use.
Immunohistochemistry (IHC)
Tumor paraffin sections were placed in a 60°C incubator for 120 min to repair the tissue sections. The Ki-67 antibody (1/100, ab15580, Abcam, UK) was added onto the tissue sections for 30 min, following by adding the anti-Rabbit IgG (ab205718) for 30 min. The sections were immersed in a methanol solution (721,964, Supelco, USA) containing 10% hydrogen peroxide (323,381, Sigma-Aldrich, USA) for 10 min, developed with Diaminobenzidine (DAB) (D3939, Sigma-Aldrich) for 3 min, counterstained with hematoxylin (H3136, Sigma-Aldrich) for 3 min. After that, the tissue sections were dehydrated and made transparent, then sealed with a neutral balsam (G8590, Solarbio, China), and ultimately analyzed by a microscope (magnification × 200).
Data Analysis
SPSS19.0 software was used for statistical analysis. Pearson was used to analyze the correlation between miR-326 and circHIPK3 expression. The correlations between the expression of circHIPK3 and clinicopathological features of the patients was analyzed by Rank sum and chi-square test. Quantitative data were expressed as mean ± standard deviation. Comparison between multiple groups was performed by one-way analysis of variance (one-way ANOVA), and comparison between two groups was performed by Student t test. P < 0.05 was a statistically significant difference.
Results
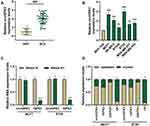
The Circular Structure of circHIPK3 Was Abundantly Expressed in the Cytoplasm and Was Frequently Up-Regulated in BCa
The data in Figure 1A and B indicated that circHIPK3 was notably upregulated in BCa tissues and cells (P < 0.01). As the expression of circHIPK3 was higher in BT20 and MCF7 cells, the two cells were used in the subsequent experiments (P < 0.001). After that, we treated the two cells with RNase R, and found that circHIPK3 had strong RNase R resistance (P < 0.001, Figure 1C). Additionally, the abundance of circHIPK3, HIPK3 and GAPDH in the cytoplasm was significantly higher than in the nucleus (Figure 1D). In addition, the correlations between the expression of circHIPK3 and clinicopathological features of BCa patients was analyzed, and the result showed that circHIPK3 expression was related to tumor size, lymph node metastasis, and TNM stage (Table 3).
 |
Table 3 The Correlations Between the Expression of circHIPK3 and Clinicopathological Features of Breast Cancer Patients |
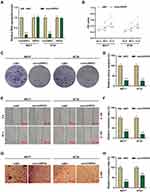
Silenced circHIPK3 Suppressed the Malignant Phenotype of BCa Cells
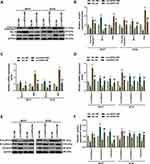
We transfected siRNA-mediated circHIPK3 into BT20 and MCF7 cells, and found that the expression of circHIPK3 was inhibited, but the expression of HIPK3 remained unchanged, indicating that the transfected sicircHIPK3 only affected circHIPK3 (P < 0.001, Figure 2A). Next, functional experiments were performed to detect the effect of circHIPK3 on the biological characteristics of BT20 and MCF7 cells. Cell viability was reduced in the sicircHIPK3 group (P < 0.01, Figure 2B). Figure 2C and D showed that the number of cell clones was memorably reduced in the sicircHIPK3 group (P < 0.001). In addition, sicircHIPK3 greatly inhibited the migration and invasion of BT20 and MCF7 cells (P < 0.001, Figure 2E–H).
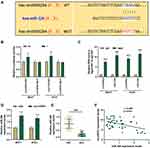
miR-326 Interacted with circHIPK3 in BCa
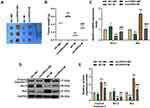
To clarify the mechanism of circHIPK3’s role in BCa, we studied its downstream targets. The website prediction results showed binding sites between miR-326 and circHIPK3 (Figure 3A). Subsequently, we co-transfected miR-326 inhibitor and circHIPK3 wild-type or mutant fluorescent reporter plasmid into BCa cells, and found that miR-326 inhibitor significantly increased the luciferase activity of circHIPK3-WT group (P < 0.01, Figure 3B). RIP assay showed that the enrichment of circHIPK3 and miR-326 in Ago2 group was higher than that in IgG group (P < 0.001, Figure 3C), indicating that circHIPK3 closely interacted with miR-326. Next, we found that the expression of miR-326 was greatly up-regulated in sicircHIPK3 (P < 0.001, Figure 3D). Moreover, the expression of miR-326 in BCa tissues was significantly down-regulated (P < 0.001, Figure 3E), and correlation analysis showed a clear negative relationship between miR-326 and circHIPK3 (r = −0.456, P = 0.001, Figure 3F).
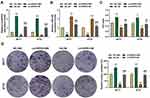
The Effect of circHIPK3 on Cell Biological Characteristics Was Reversed by miR-326 in BCa Cells
To verify whether circHIPK3 regulated the malignant transformation of BCa cells through sponging miR-326, circHIPK3 overexpression plasmid and miR-326 mimic were successfully transfected into BT20 and MCF7 cells. As shown in Figure 4A, the expression of circHIPK3 was up-regulated by circHIPK3 and down-regulated by miR-326 mimic (P < 0.001, Figure 4A). The expression of miR-326 was up-regulated by miR-326 mimic and down-regulated by circHIPK3, and miR-326 mimic could relieve the inhibition of miR-326 by circHIPK3 (P < 0.001, Figure 4B). Next, we observed that miR-326 mimic inhibited cell viability and proliferation, migration and invasion, while circHIPK3 had the opposite effect, and the promotive effect of circHIPK3 on the aforementioned effect was partially reversed by miR-326 mimic (P < 0.05, Figure 4C and D, 5A–D). Furthermore, circHIPK3 slowed down cell apoptosis and miR-326 mimic-induced cell apoptosis, and the circHIPK3+M group partially overturned miR-326 mimic or circHIPK3 on cell apoptosis (P < 0.001, Figure 5E and F).
miR-326 Mimic Partially Offset the Effects of circHIPK3 on Apoptosis and EMT-Related Proteins
The overexpression of circHIPK3 up-regulated the expression of Bcl-2, inhibited the expression of cleaved Caspase-3 and Bax, while the up-regulated miR-326 showed the opposite effect. In addition, miR-326 mimic partially counteracted the effect of circHIPK3 (P < 0.05, Figure 6A–C). EMT plays an important role in the malignant progression of tumors; therefore, we detected the expression of EMT-related genes. Transfection of circHIPK3 overexpression plasmid promoted in the expression of N-Cadherin and Vimentin and suppressed the expressions of E-Cadherin, and the effect of circHIPK3 in EMT-related genes was partially reversed by miR-326 mimic (P < 0.05, Figure 6D–F).
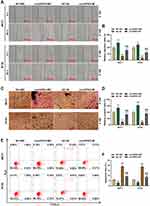
CircHIPK3 Affected Tumor Growth and Apoptosis in Mice by Sponging miR-326
According to Figure 7A and B, compared with NC+MC, tumor size and weight were significantly reduced in NC+M group but significantly increased in circHIPK3+MC group. Moreover, circHIPK3+M group partially overturned the effects of miR-326 mimic or circHIPK3 on the growth of transplanted tumors (P < 0.001). Then, we examined the expression of apoptosis genes in tumor tissues, and found that overexpressed circHIPK3 up-regulated the expression of Bcl-2, inhibited the expressions of cleaved Caspase-3 and Bax, while miR-326 mimic had the opposite regulatory effect. Importantly, the regulatory function of overexpressed circHIPK3 on apoptosis proteins was reversed by miR-326 mimic (P < 0.05, Figure 7C–E).
Compared with NC+MC, circHIPK3 expression was significantly up-regulated in circHIPK3+MC group and significantly down-regulated in NC+M group. circHIPK3 +M group partially overturned the function of miR-326 mimic or circHIPK3 (P < 0.001, Figure 8A). Inversely, miR-326 was inhibited by overexpressed circHIPK3 and promoted by miR-326 mimic, and miR-326 mimic partially relieved the effect of overexpressed circHIPK3 (P < 0.001, Figure 8B). In addition, overexpressed circHIPK3 promoted the level of Ki-67, while miR-326 mimic inhibited the level of Ki-67. CircHIPK3 + M group partially overturned the effect of circHIPK3 + MC group or NC + M group on Ki-67 level (Figure 8C and D).
Discussion
The continuous in-depth research on the regulatory mechanism of cancer has the research direction has gradually shifted from coding RNAs to non-coding RNAs.19 The function and mechanism of non-coding RNAs in cancer have been widely recognized in the past ten years.20,21 CircRNAs play important roles in the occurrence and development of BCa, and due to its high conservation and tissue specificity, it has the potential to become tumor markers for screening BCa.9 In this study, we found that circHIPK3 downregulation attenuated BCa cell proliferation, migration, invasion and EMT, and decelerated BCa tumor growth via upregulating miR-326.
CircRNA has highly stable, conserved and tissue-specific expression, and is a potential cancer biomarker.22 In this study, it was confirmed that circHIPK3 was not easily degraded by RNase R and was abundantly expressed in the cytoplasm. Differentially expressed circRNA has been widely detected in cancer cells. For instance, Chen et al found that the expression levels of circRNAs in normal human endometrial cancer tissues were low.23 Researchers screened 235 differentially expressed circRNAs in BCa, and found that the differentially expressed circTADA2As inhibited the malignant phenotype of BCa cells.24 CircHIPK3 was abnormally expressed in BCa samples and cell lines, suggesting that circHIPK3 may be a new target for the diagnosis of BCa.
Next, we studied the effect of circHIPK3 on cell biological functions by exogenously interfering with circHIPK3 in vitro and in vivo, and found that circHIPK3 acted as an oncogene in BCa to promote cell proliferation, EMT and reduce apoptosis, and accelerate tumorigenesis. Our study was similar to that of Cai et al.25 Their study in prostate cancer showed that circHIPK3 regulates the expression of ADAM17 through miR-338-3p and can accelerate the malignant progression of prostate cancer.25 The latest research confirmed the findings of our research,26 except that in their study the effect of circHIPK3 was realized via miR-193a/HMGB1/PI3K/AKT axis.26 However, we found that miR-326 was also targeted by circHIPK3 and the two were negatively correlated.
Equally important, circRNA molecules are widely involved in the translation and expression of proteins, and they also have the functions of miRNA sponge molecules and RNA binding protein.27 The overexpression of CDRIas can promote the proliferation, invasion and migration of cervical cancer cells by inhibiting the activity of miR-7 and up-regulating the expression of FAK.28 We found that up-regulation of miR-326 can counteract the effect of circHIPK3 on promoting the malignant progression of BCa, suggesting that circHIPK3 can promote tumorigenesis and metastasis in BCa by inhibiting the expression of miR-326. A large amount of data showed that miR-326 was a key mediator in the pathogenesis of many tumors and can be a potential target for new cancer therapies.29 For example, miR-326 expression is down-regulated in colorectal cancer tissues, while that of its downstream target NOB1 is up-regulated; miR-326 can inhibit the expression of NOB1 to reduce the proliferation and metastasis of colorectal cancer cells.30
Studies have confirmed that miRNA expression profiles are different in BCa and normal breast tissues, suggesting that miRNA and BCa are closely related.31 Theoretically, in hepatocellular carcinoma cell lines and tissues, the expression of miR-326 is significantly down-regulated; miR-326 affects cell cycle progression and promotes apoptosis through PDK1 to reduce the growth of hepatocellular carcinoma cells and EMT phenotype and inhibit HCC cell invasion.32 It has been reported that circ-TFF1 contributes to breast cancer progression through targeting miR-326/TFF1 signaling.33 miR-326 functions as a tumor suppressor in breast cancer by inhibiting ErbB/PI3K pathway.34 miR-326 is low-expressed in human breast cancer tissues and cell lines, and miR-326 inhibited breast cancer cell proliferation, migration and invasion via targeting SOX12.35 In our study, the expression of miR-326 was down-regulated, and miR-326 also inhibited cell growth, suggesting that miR-326 may be a reliable therapeutic target. However, there is limitation of this study, that is, the effect of circHIPK3 in multiple models of each of the breast cancer subtypes should be further studied. In addition, MCF7 and BT20 cell lines have the higher levels of circHIPK3 in comparison with the other BCa cell lines, this needs more experiments to explore this problem.
Conclusion
To conclude, this research has explained the importance of circHIPK3 in BCa. Mechanistically, circHIPK3 affects the apoptosis pathway and EMT phenotype by targeting miR-326 to promote the growth and metastasis of BCa tumors.
Funding
This work was supported by the Natural Science Foundation of China [81001187].
Disclosure
The authors declare no conflicts of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Peart O. Breast intervention and breast cancer treatment options. Radiol Technol. 2015;86(5):
3. Brook N, Brook E, Dharmarajan A, Dass CR, Chan A. Breast cancer bone metastases: pathogenesis and therapeutic targets. Int J Biochem Cell Biol. 2018;96:63–78.
4. Sun YS, Zhao Z, Yang ZN, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13(11):1387–1397. doi:10.7150/ijbs.21635
5. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
6. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
7. Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10(2):592–604.
8. Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123(13):2113–2118.
9. Li Z, Chen Z, Hu G, Jiang Y. Roles of circular RNA in breast cancer: present and future. Am J Transl Res. 2019;11(7):3945–3954.
10. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi:10.1038/ncomms11215
11. Wu J, Jiang Z, Chen C, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi:10.1016/j.canlet.2018.05.033
12. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
13. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO, Moran JV. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi:10.1371/journal.pgen.1003777
14. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
15. Liu N, Zhang J, Zhang L-Y, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718. doi:10.26355/eurrev_201806_15250
16. Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018;503(3):1570–1574. doi:10.1016/j.bbrc.2018.07.081
17. Zhang JX, Lu J, Xie H, et al. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10(3):182. doi:10.1038/s41419-019-1430-7
18. Wang ST, Liu LB, Li XM, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma. 2019;66(2):232–239. doi:10.4149/neo_2018_180710N460
19. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi:10.1038/nrd.2016.117
20. Hulshoff MS, Del Monte-Nieto G, Kovacic J, Krenning G. Non-coding RNA in endothelial-to-mesenchymal transition. Cardiovasc Res. 2019;115(12):1716–1731. doi:10.1093/cvr/cvz211
21. Klinge CM. Non-coding RNAs in breast cancer: intracellular and intercellular communication. Noncoding RNA. 2018;4(4). doi:10.3390/ncrna4040040
22. Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630–3638. doi:10.1111/cas.14211
23. Chen BJ, Byrne FL, Takenaka K, et al. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2018;9(5):5786–5796. doi:10.18632/oncotarget.23534
24. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. doi:10.1038/s41419-019-1382-y
25. Cai C, Zhi Y, Wang K, et al. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi:10.2147/OTT.S196931
26. Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ, Wang GS. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac Cancer. 2020;11(9):2660–2671. doi:10.1111/1759-7714.13603
27. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi:10.1016/j.pharmthera.2018.01.010
28. Lee BY, Timpson P, Horvath LG, Daly RJ. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132–149.
29. Pan YJ, Wan J, Wang CB. MiR-326: promising biomarker for cancer. Cancer Manag Res. 2019;11:10411–10418. doi:10.2147/CMAR.S223875
30. Wu L, Hui H, Wang LJ, Wang H, Liu QF, Han SX. MicroRNA-326 functions as a tumor suppressor in colorectal cancer by targeting the nin one binding protein. Oncol Rep. 2015;33(5):2309–2318. doi:10.3892/or.2015.3840
31. Loh HY, Norman BP, Lai KS, Rahman N, Alitheen NBM, Osman MA. The regulatory role of MicroRNAs in breast cancer. Int J Mol Sci. 2019;20:19. doi:10.3390/ijms20194940
32. Mo Y, He L, Lai Z, et al. Gold nano-particles (AuNPs) carrying miR-326 targets PDK1/AKT/c-myc axis in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol. 2019;47(1):2830–2837. doi:10.1080/21691401.2018.1489266
33. Pan G, Mao A, Liu J, Lu J, Ding J, Liu W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53(2):e12720. doi:10.1111/cpr.12720
34. Ghaemi Z, Soltani BM, Mowla SJ. MicroRNA-326 functions as a tumor suppressor in breast cancer by targeting ErbB/PI3K signaling pathway. Front Oncol. 2019;9:653. doi:10.3389/fonc.2019.00653
35. Du Y, Shen L, Zhang W, et al. Functional analyses of microRNA-326 in breast cancer development. Biosci Rep. 2019;39:7. doi:10.1042/BSR20190787
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.