Back to Journals » OncoTargets and Therapy » Volume 13
CircHIPK3 Facilitates the G2/M Transition in Prostate Cancer Cells by Sponging miR-338-3p
Authors Liu F, Fan Y, Ou L, Li T, Fan J, Duan L, Yang J, Luo C , Wu X
Received 15 December 2019
Accepted for publication 1 May 2020
Published 22 May 2020 Volume 2020:13 Pages 4545—4558
DOI https://doi.org/10.2147/OTT.S242482
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Fengchun Liu,1 Yanru Fan,1 Liping Ou,1 Ting Li,1 Jiaxin Fan,1 Limei Duan,1 Jinxiao Yang,1 Chunli Luo,1 Xiaohou Wu2
1Department of Laboratory Diagnosis, Chongqing Medical University, Yuzhong, Chongqing 408000, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Chongqing Medical University, Yuzhong, Chongqing 408000, People’s Republic of China
Correspondence: Xiaohou Wu
Department of Urology, The First Affiliated Hospital of Chongqing Medical University, No. 1 Medical College Road, Yuzhong, Chongqing 408000, People’s Republic of China
Email [email protected]
Chunli Luo
Department of Laboratory Diagnosis, Chongqing Medical University, No. 1 Medical College Road, Yuzhong, Chongqing 408000, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) play a crucial role in gene expression regulation. CircHIPK3 is a circRNA derived from Exon 2 of HIPK3 gene and its role in prostate cancer (PCa) is still unclear.
Methods: CCK8 assays, flow cytometry and colony formation assays were performed to assess the effects of circHIPK3 in PCa cells. Bioinformatics analysis, RNA pull-down assay, RNA immunoprecipitation assay (RIP), and luciferase activity assay were performed to dissect the mechanism underlying circHIPK3-mediated G2/M transition in PCa cells.
Results: CircHIPK3 expression was upregulated in PCa cells and prostate cancer tissues. Overexpression of circHIPK3 or circHIPK3 silencing altered PCa viability, proliferation and apoptosis in vitro. CircHIPK3 could sponge miR-338-3p and inhibit its activity, resulting in increased expression of Cdc25B and Cdc2 in vitro.
Conclusion: CircHIPK3 promotes G2/M transition and induces PCa cell proliferation by sponging miR-338-3p and increasing the expression of Cdc25B and Cdc2. CircHIPK3 may play an oncogenic role in PCa.
Keywords: prostate cancer, circHIPK3, miR-338-3p, Cdc25B, G2/M transition
Introduction
Prostate cancer (PCa) is one of the most commonly diagnosed cancers among men worldwide.1 Once the tumor has migrated out of the gland, an incurable metastatic disease may inevitably occur.2–4
Circular RNA (circRNA) is an endogenous non-coding RNA.4 Unlike linear RNA terminated with a 5ʹ cap and a 3ʹ tail, circRNA forms a covalent closed-loop structure.4,5 At present, molecular purification methods combined with next-generation RNA sequencing have led to an in-depth understanding of circRNA.6 A mass of circRNAs are conserved throughout species with specific expression at the tissue/developmental stage, and are stable and resistant to RNaseR.7,8 CircRNAs provide new insights into the pathogenesis of diseases.9
MicroRNAs (miRNAs) are endogenous ~23 nt RNAs that play important gene-regulatory roles in animals by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression.10 Gain-of-function and loss-of-function experiments, in combination with target prediction analyses, have demonstrated that microRNAs can affect different steps of the tumorigenesis and progression of various cancers, including PCa.11,12
CircHIPK3 is derived from exon 2 of the HIPK3 gene. It has been shown that circHIPK3 sponges miR-124 and suppresses miR-124 activity.13 In colorectal cancer, circHIPK3 may have considerable potential as a prognostic biomarker, and support the notion that therapeutic targeting of the c-Myb/circHIPK3/miR-7 axis may be a promising treatment approach for patients.14 CircHIPK3 inhibited the expression of heparanase (HPSE) in bladder cancer by sponging miR-558.15 CircHIPK3 plays an important role in tumors and diabetic retinopathy by sponging miR-30a.16 Recently, it has been reported that circHIPK3 was upregulated in PCa tissues and correlated with proliferation and invasion.17,18 Until now, little known about the potential role of circHIPK3 in cell cycle of prostate cancer cells, and its molecular mechanism still remains elusive.
In this study, we reported that circHIPK3 is an oncogene that is upregulated in PCa. Importantly, we found that circHIPK3 knockdown markedly arrested the G2/M transition in PCa cells. Furthermore, circHIPK3 could sponge miR-338-3p to inhibit miR-338-3p activity, thereby leading to increased Cdc25B and Cdc2 expression. Our findings may reveal a novel mechanism of circHIPK3 in the development of PCa and provide a new perspective for the treatment of PCa.
Materials and Methods
Tissue Samples
PCa tissues and benign prostatic hyperplasia (BPH) tissues were surgically removed from 45 patients with PCa and 25 patients with BPH, respectively, and frozen in liquid nitrogen immediately. Clinical pathology data were collected from patients. Patients volunteered to participate in the study and signed written informed consent. This study was approved by the Ethics Committee of Chongqing Medical University (Approval No. 20176503) and was implemented following the Helsinki Declaration.
Cell Culture and Transfection
Human prostate cell lines (LNCaP, DU145, PC3 and 22RV1) and Human prostate epithelial cell line RWPE-1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). 22RV1, LNCaP and DU145 cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin (PS; Sangon, Shanghai, China). PC3 cells were cultured in F-12 medium containing 10% FBS and 1% PS. RWPE-1 cells were cultured in Keratinocyte-SFM (K-sfm; Gibco) medium with Bovine Pituitary Extract (20–30 μg/mL) and rEGF (0.1–0.2 ng/mL) containing 10% FBS and 1% PS. The cells were placed in an incubator at 37°C with 5% CO2. The cells were transfected at approximately 60% confluence with synthesized small interfering RNAs (siRNAs) (100 nmol/L; GenePharma, Shanghai, China) targeting circHIPK3 using EndoFectin™-Max reagent (GeneCopoeia Inc., Rockville, MD, USA).
Quantitative PCR (qPCR)
Total RNA was extracted from cells and clinical samples with TRIzol (Life Technologies, Carlsbad, CA, USA). The nuclear and cytoplasmic fractions were extracted with NE-PER nuclear and cytoplasmic extraction kits (Thermo Scientific, Waltham, MA, USA). cDNA was synthesized from total RNA using the PrimeScript RT Master Mix (Takara, Dalian, China), and qPCR was carried out using TB Green Premix Ex Taq II (Takara). Stem-loop RT-PCR was used to detect the amount of miRNA. The primers are listed in Table S1.
RNA Fluorescence in situ Hybridization (FISH)
The Cy3-labelled circHIPK3 probe and FAM-labelled miR-338-3p probe were designed and synthesized by GenePharma (Shanghai, China). The probe sequences are listed in Table S1. FISH was carried out using the kit (GenePharma). The images were gained on a ZEISS 800 laser scanning confocal microscope (ZEISS, Jena, Germany).
CircHIPK3 Plasmids Construction
To construct circHIPK3 over-expression plasmids, human circHIPK3 cDNA was synthesized by TSINGKE (Nanjing, China) and cloned into pLCDH-ciR vector (Geenseed Biotech Co, Guangzhou, China). The pLCDH-ciR vector contained a front circular frame and a back circular frame.
Cell Counting Kit-8 Proliferation Assay
The proliferative capacity of LNCaP and DU145 cells was evaluated by CCK8 (Dojindo Laboratories, Kumamoto, Japan) analysis. The results were read on an automatic microplate reader (Synergy 4; BioTek, Winooski, VT, USA) at 450 nm spectrophotometry.
Colony Formation Assay
PCa cells were seeded in 6-well plates at a density of 500 cells/well and cultured for 2 weeks. The colonies were fixed with methanol and stained with crystal violet. Finally, colonies containing ≥50 cells were counted under a microscope (Olympus, Toyko, Japan).
Flow Cytometry Analysis
Flow cytometry was applied to detect cell cycle and apoptosis. PCa cells were stained with propidium iodide (PI) (Beyotime, C1052, Shanghai, China). Cells were washed and resuspended in cold phosphate buffered saline (PBS) and incubated in ice-cold 70% ethanol for 4 h. The cells were then centrifuged at 1500 rpm for 10 min and resuspended in propidium iodide (PI) master mix at a density of 5ⅹ105 cells/mL and incubated at 37°Cfor 30 min before analysis with flow cytometry as described. For apoptosis assays, cells were harvested as before and double stained with fluorescein isothiocyanate (FITC)-conjugated Annexin V and PI. Apoptotic cells were detected using Annexin V-FITC/PI staining kit (BestBio, BB-4101-3, Shanghai, China).
Dual-Luciferase Reporter Assay
293T cells were seeded in six-well plates, and then co-transfected with wild-type or mutant circHIPK3 or Cdc25B-3ʹUTR reporter plasmids, and miR-338-3p mimics or miR-NC. Luciferase assay was performed using TransDetect® dual-luciferase reporter assay kit (TransGen, Beijing, China) and Dual-luciferase reporter assay system (Promega, WI, USA). The ratio of firefly fluorescence to Renilla fluorescence was taken as relative luciferase activity.
RNA Immunoprecipitation Assay (RIP)
RIP was carried out using the BersinBioTM RIP kit (BersinBio, Guangzhou, China) according to the manufacturer’s instructions. The miR-338-3p mimic or miR-NC was transfected into cells. After 48 h, 107 cells were collected and lysed in 850 μL RNA lysis buffer. The cell lysates were incubated with 5 μg Rabbit anti-Argonaute2 (Ago2) (2897, 1:50, CST, San Antonio, TX, USA) or negative control IgG antibody-conjugated magnetic beads at 4°C for 4 h. A total of 40 μL of protein A-Sepharose was added to each sample, and the mixtures were incubated at 4°C for 4 h. The pellets were washed 3×with PBS and resuspended in 0.5 mL TriReagent. Finally, coprecipitated RNA was detected by qPCR.
RNA Pull-Down
Pull-down assay was performed following the manufacturer’s instructions of BersinBioTM RNA pull down kit. Biotinylated circHIPK3 probe was designed and synthesized by BersinBio. In brief, 107 cells were washed in cold PBS, lysed in 750 μL RIP buffer, and incubated with 2 μg biotinylated circHIPK3 probes, at room temperature for 2 h. A total of 50 μL washed StreptavidinC1 magnetic beads were added to each binding reaction and further incubated at room temperature for another hour. The beads were washed briefly with RIP buffer for five times. The eluted RNA in the pull-down materials were analyzed by qPCR. The circHIPK3 probes sequences are listed in Table S1.
Western Blot Analysis
Cell lysates were prepared with RIPA buffer, and the protein concentration was measured using bicinchoninic acid (BCA) protein assay kit (Beyotime). Then, equal amounts of protein were resolved by 10% SDS-PAGE analysis and electrotransferred onto an NC membrane (Millipore, Schwalbach, Germany), then blocked with 5% skim milk powder and incubated with primary antibody at 4°C overnight. The primary antibodies were used: Polyclonal Goat anti-Cdc25B (AF1649, 1:6000, R&D, CA, USA), Mouse anti-Cdc2 (9116, 1:1000, CST), Rabbit anti-p-Cdc2 (4539, 1:1000, CST), and Rabbit anti-β-actin (3700, 1:1000, CST); HRP-conjugated secondary goat anti-mouse (ZB2305, 1:20,000, Zsbio, Shanghai, China), goat anti-rabbit (ZB2301) and rabbit anti-goat antibodies (ZB2306). Immunoreactive bands were detected by using the Super SignalTM West Femto maximum sensitivity substrate (ThermoFisher). The images were acquired using a protein imaging system (Proteinsimple, lourCherm FC3, China).
Immunohistochemistry (IHC) Staining
IHC staining was carried out according to the manufacturer of SPlink Detection Kits (Zsbio). The slides were placed in 3%H2O2 for 10 min at room temperature. After washing with PBS, they were blocked with Reagent A at 37°C for 30 min. Polyclonal Goat anti-Cdc25B (R&D) were diluted 1:200, incubated with the slides overnight at 4°C. The next day, the slides were washed with PBS and incubated with Reagent B at 37°C for 30 min, then incubated with Reagent C at 37°C for 30 min. After washing, DAB (Zsbio) staining was performed under the microscope. After rinsing with distilled water, the slides were routinely counterstained, dehydrated, cleared, and mounted. Each sample was incubated with an isotypic antibody dilution under the same experimental conditions as the negative control. Images were acquired using an optical microscope (Olympus, BX53) and analyzed with image-processing software (ImageJ v 1.48). The degree of positivity was determined according to the percentage of positive tumor cells.
Statistical Analysis
The statistical analysis was performed using SPSS 19.0 (IBM, SPSS, Chicago, IL, USA). The data are presented as the mean ± standard error (SEM) or standard deviation (SD). The differences between groups were assessed by Student’s t-test, one-way ANOVA or two-way ANOVA. P values <0.05 were considered to be significant.
Results
CircHIPK3 Is Upregulated in PCa and Predominantly Localized in the Cytoplasm
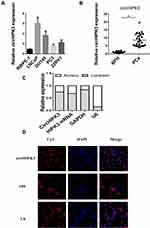
qPCR was used to detect the expression of circHIPK3 in PCa tissues and PCa cell lines. The expression levels of circHIPK3 in LNCaP and DU145 cells were upregulated by about 6-fold and 4-fold compared with the normal prostate epithelial cells (RWPE-1) (Figure 1A). CircHIPK3 was upregulated approximately 15-fold in PCa tissues compared to BPH tissues (Figure 1B). PCa patients with high expression of circHIPK3 had higher Gleason scores than patients with low expression (Table 1). Next, we evaluated the localization of circHIPK3 by qPCR and FISH assays. The results showed that circHIPK3 was predominantly localized in the cytoplasm of LNCaP cells (Figure 1C and D).
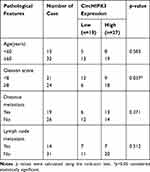 |
Table 1 Correlation of CircHIPK3 Expression with Clinical Features of Prostate Cancer (n=45) |
Silencing of CircHIPK3 Inhibits LNCaP and DU145 Cells Proliferation and Induces Apoptosis
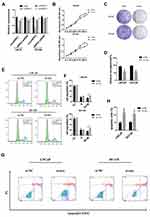
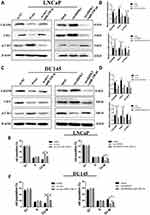
In order to explore the function of circHIPK3, three siRNAs were designed against circHIPK3 junction site. After transfecting the three siRNAs into LNCaP and DU145 cells, only si-circHIPK3-1 significantly reduced the expression of circHIPK3 without affecting the expression level of homologous HIPK3 mRNA (Figure 2A). Therefore, si-circHIPK3-1 was chosen for the following experiment. CCK8 assay showed that the cell proliferation ability decreased in LNCaP and DU145 cells with circHIPK3 knockdown (Figure 2B). The colony formation assay showed that the colony-forming capacity of LNCaP and DU145 cells was significantly suppressed with circHIPK3 knockdown (Figure 2C and D). Flow cytometry showed that silencing of circHIPK3 arrested cell cycle at the G2/M phase (Figure 2E and F), and increased apoptotic cells in LNCaP and DU145 cells (Figure 2G and H). Taken together, these data suggest that circHIPK3 silencing inhibits PCa progression.
CircHIPK3 Sponges miR-338-3p in LNCaP and DU145 Cells
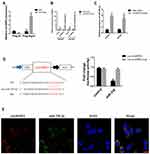
CircHIPK3 functions as a “miRNA sponge” in HEK-293T cells.13 To detect whether circHIPK3 could sponge miRNA in PCa cells, we used the CircInteractome Database to select candidate miRNAs (https://circinteractome.nia.nih.gov/index.html).19 We found that circHIPK3 possessed many binding sites for miRNAs, including miR-338-3p. Next, we tested whether circHIPK3 could bind miR-338-3p. qPCR analysis showed that circHIPK3 was pulled down from LNCaP cells overexpressing Flag-Ago2 (Figure 3A), suggesting that circHIPK3 can bind to Ago2. A biotin-labelled circHIPK3 probe was used to pull-down circHIPK3 in both LNCaP and DU145 cells, and circHIPK3 overexpression enhanced the pull-down efficiency (Figure 3B). qPCR analysis revealed that miR-338-3p was abundantly pulled down by the circHIPK3 probe in both LNCaP and DU145 cells (Figure 3C). Furthermore, luciferase reporter assay showed that relative luciferase activity of the vector containing the unmutated miR-338-3p binding site was reduced by about 26%. However, the luciferase activity of the mutated miR-338-3p binding site did not change (Figure 3D). Moreover, FISH assay showed that circHIPK3 and miR-338-3p were co-localized in the cytoplasm in LNCaP cells (Figure 3E). Collectively, these results suggested that circHIPK3 can bind to miR-338-3p in LNCaP and DU145 cells.
Overexpression of CircHIPK3 Partially Reverses miR-338-3p Induced Repression of LNCaP and DU145 Cells Cycle Progression
miR-338-3p may participate in the progression of many human tumors, including PCa.20 Overexpression of miR-338-3p facilitated the G2/M transition and induced LNCaP and DU145 cells apoptosis, resembling the function of circHIPK3 silencing. To explore whether circHIPK3 has oncogenic role by sponging miR-338-3p, we co-transfected si-circHIPK3 or circHIPK3 vector and miR-338-3p mimic into LNCaP and DU145 cells. The results showed that the cell cycle was arrested at G2/M phase and apoptotic cells increased in the miR-338-3p+si-circHIPK3 group. However, these effects were partially reversed in circHIPK3 vector + miR-338-3p group (Figure 4A–D). These results demonstrate that overexpression of circHIPK3 reverses miR-338-3p-induced repression of PCa cell cycle progression.
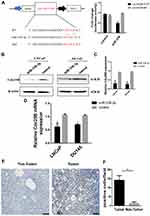
Cdc25B Is a Direct Target of miR-338-3p in LNCaP and DU145 Cells
Based on StarBase prediction (http://starbase.sysu.edu.cn/), we identified several targets of miR-338-3p, including Cdc25B. To further identify whether miR-338-3p interacts with the 3ʹ-UTR of Cdc25B in PCa cells, we performed luciferase reporter assay. Co-transfection of miR-338-3p mimics and LUC-Cdc25B or LUC-Cdc25B mutant into 293T cells showed that the activity of luciferase was decreased by around 50% in the LUC-Cdc25B group compared with the negative control, but the luciferase activity of the LUC-Cdc25B-mut group was not changed (Figure 5A). Furthermore, the gain and loss of function assays showed that overexpression of miR-338-3p inhibited the expression of Cdc25B at both protein and mRNA level(Figure 5B–D). At the same time, the results of IHC showed that Cdc25B was highly expressed in PCa tissues (Figure 5E and F). These data indicate that Cdc25B is a direct target of miR-338-3p in LNCaP and DU145 cells.
CircHIPK3 Facilitates LNCaP and DU145 Cells G2/M Transition via CircHIPK3/miR-338-3p/Cdc25B Axis
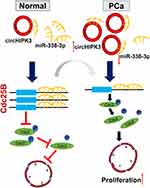
Finally, we investigated whether circHIPK3 exerted tumor-promoting effect by increasing the expression of the oncogenes targeted by miR-338-3p. The results showed that the expression of Cdc25B and Cdc2 decreased while the expression of p-Cdc2 increased in LNCaP cells co-transfected with si-circHIPK3 and miR-338-3p inhibitor compared with cells transfected with si-circHIPK3 alone(Figure 6A–D). Similarly, the expression of Cdc25B and Cdc2 decreased while the expression of p-Cdc2 increased in LNCaP and DU145 cells co-transfected with miR-338-3p mimics and circHIPK3 vector compared with cells transfected with circHIPK3 vector alone(Figure 6A–D). More LNCaP and DU145cells were arrested at the G2/M phase in the si-circHIPK3+miR-338-3p inhibitor group compared to the si-circHIPK3 group. Moreover, the co-transfection of circHIPK3 vector and miR-338-3p mimic facilitated the G2/M transition of LNCaP and DU145 cells compared with circHIPK3 transfection(Figure 6E and F). Collectively, these results indicate that circHIPK3 facilitates LNCaP and DU145 cells G2/M transition via sponging miR-338-3p and subsequent upregulation of Cdc25B(Figure 7).
Discussion
Recently, numerous circRNAs have been identified by next-generation sequencing technology, most of which have highly stable expression.5 An increasing amount of studies showed that circRNA has a vital role in tumorigenesis.21,22 The role of circHIPK3 in different diseases has been reported.13–16 CircHIPK3 and its target miRNAs are also involved in regulating proliferation and invasion of PCa.17,18 In our study, we also found that circHIPK3 is upregulated in PCa tissues comparing with BPH, and the high expression of circHIPK3 is related to the patients’ Gleason scores. However, the molecular mechanism of circHIPK3 in cell cycle of prostate cancer cells has not yet been elucidated clearly. In this study, we focused on the role of circHIPK3 in cell cycle of prostate cancer cells, and found that it can facilitate the G2/M transition in prostate cancer cells.
It is well known that circRNAs contain miRNA response elements (MREs) that could function as miRNA sponges.23,24 Previous studies have reported that circRNAs regulate the expression of target genes in a variety of human diseases, including cancer, by acting as a sponge for miRNAs. For example, circMTO1 suppressed HCC progression by promoting p21 expression through sponging miR-9.25 Additionally, circHIPK3 could efficiently inhibit the expression of heparanase in human bladder cancer by sponging miR-558.15 In this study, bioinformatics analyses, RNA pull down and dual-luciferase reporter assay showed that circHIPK3 could sponge miR-338-3p. FISH assay demonstrated that circHIPK3 and miR-338-3p were colocalized in the cytoplasm of LNCaP cells. miR-338-3p was downregulated in some cancers, including gastric cancer, ovarian cancer, breast cancer, and PCa.20,26-29 Overexpression of miR-338-3p in PCa cells repressed cell migration and invasion.17,30 We discovered that overexpression of circHIPK3 could reverse miR-338-3p-induced induced apoptosis and cell cycle arrest at G2/M phase in LNCaP and DU145 cells, firstly. Knockdown circHIPK3 has got the opposite results. So we assumed that circHIPK3 and miR-338-3p may affect the progression of PCa through another pathway, not only circHIPK3/miR-338-3p/ADAM17.17
Cdc25B is a key regulator of cell cycle by activating CDK complexes.31 Bioinformatics analysis predicted that miR-338-3p could directly target Cdc25B. The interaction was confirmed by the dual-luciferase reporter assay. It has been shown that Cdc25B expression was upregulated in PCa.32 In this study, we found that Cdc25B level significantly decreased in LNCaP and DU145 cells transfected with miR-338-3p mimics. During the M/G1 phase of the cell-cycle, Cdc25B is nuclear and moves to the cytoplasm during S and G2.33 Our study focuses on the G2/M phase that Cdc25B mainly localized in the cytoplasm during this phase. Cdc25B dephosphorylates at Thr14 and Tyr15 in the cyclin-dependent kinase Cdc2 to make it active. This dephosphorylation is necessary for Cdc2 to enter into mitosis.31 Cdc2 is a key player in the transition of G2/M in mitotic cells.34 In our study, silencing circHIPK3 not only decreased the expression of Cdc25B and Cdc2 as well as increased the expression of p-Cdc2 but also arrested the cell cycle at the G2/M phase in LNCaP and DU145 cells. In contrast, the overexpression of circHIPK3 upregulated the expression of Cdc25B and Cdc2 as well as downregulated the expression of p-Cdc2 and facilitated G2/M transition in LNCaP and DU145 cells. These effects could be reversed by miR-338-3p inhibitor or miR-338-3p mimic, respectively. Collectively, these findings indicate that circHIPK3 regulates the expression of Cdc25B and affect cell cycle of LNCaP and DU145 cells via sponging miR-338-3p.
So our study certified a new assumption that circHIPK3 acted as miR-338-3p sponge to adjust the expression of Cdc25B. The result may provide a new perspective for treatment of PCa.
Conclusion
In summary, we found that circHIPK3 was upregulated in PCa tissues and cells and played an oncogenic role in PCa. Mechanistically, circHIPK3 sponges miR-338-3p and inhibits its activity, resulting in increased Cdc25B expression and enhanced G2/M transition in LNCaP and DU145 cells.
Acknowledgments
We thank the staff who help collect patient tissue samples in the Department of Urology, the First Affiliated Hospital of Chongqing Medical University.
Disclosure
The authors declare that they have no competing interest.
References
1. Mohler JL, Armstrong AJ, Bahnson RR, et al. Prostate cancer, version 1.2016. J Natl Compr Canc Netw. 2016;14(1):19–30. doi:10.6004/jnccn.2016.0004
2. Jackson WC, Suresh K, Tumati V, et al. Intermediate endpoints after postprostatectomy radiotherapy: 5-year distant metastasis to predict overall survival. Eur Urol. 2018;74(4):413–419. doi:10.1016/j.eururo.2017.12.023
3. Xie W, Regan MM, Buyse M, et al. Metastasis-free survival is a strong surrogate of overall survival in localized prostate cancer. J Clin Oncol. 2017;35(27):3097–3104. doi:10.1200/JCO.2017.73.9987
4. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi:10.1261/rna.035667.112
5. Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi:10.1371/journal.pone.0030733
6. Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43(5):2525–2534. doi:10.1093/nar/gkv115
7. Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife. 2015;4:e07540. doi:10.7554/eLife.07540
8. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi:10.1038/nrm.2015.32
9. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881 e813. doi:10.1016/j.cell.2018.12.021
10. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
11. Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169(5):1812–1820. doi:10.2353/ajpath.2006.060480
12. He S, Zhu L, Liu F, et al. Functions of the Vasa gene in Schistosoma japonicum as assessed by RNA interference. Gene. 2018;638:13–19. doi:10.1016/j.gene.2017.09.054
13. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi:10.1038/ncomms11215
14. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
15. Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi:10.15252/embr.201643581
16. Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi:10.1161/CIRCULATIONAHA.117.029004
17. Cai C, Zhi Y, Wang K, et al. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi:10.2147/OTT.S196931
18. Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. 2019;11:1415–1423. doi:10.2147/CMAR.S190669
19. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
20. Zhang P, Shao G, Lin X, Liu Y, Yang Z. MiR-338-3p inhibits the growth and invasion of non-small cell lung cancer cells by targeting IRS2. Am J Cancer Res. 2017;7(1):53–63.
21. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
22. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
23. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
24. Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–2246. doi:10.1093/bioinformatics/btu257
25. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi:10.1002/hep.29270
26. Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8(7):e2928. doi:10.1038/cddis.2017.325
27. Nie H, Li J, Yang XM, et al. Mineralocorticoid receptor suppresses cancer progression and the warburg effect by modulating the miR-338-3p-PKLR axis in hepatocellular carcinoma. Hepatology. 2015;62(4):1145–1159. doi:10.1002/hep.27940
28. Sun F, Yu M, Yu J, et al. miR-338-3p functions as a tumor suppressor in gastric cancer by targeting PTP1B. Cell Death Dis. 2018;9(5):522. doi:10.1038/s41419-018-0611-0
29. Zhang Y, Shi B, Chen J, Hu L, Zhao C. MiR-338-3p targets pyruvate kinase M2 and affects cell proliferation and metabolism of ovarian cancer. Am J Transl Res. 2016;8(7):3266–3273.
30. Wang Y, Qin H. miR-338-3p targets RAB23 and suppresses tumorigenicity of prostate cancer cells. Am J Cancer Res. 2018;8(12):2564–2574.
31. Sur S, Agrawal DK. Phosphatases and kinases regulating CDC25 activity in the cell cycle: clinical implications of CDC25 overexpression and potential treatment strategies. Mol Cell Biochem. 2016;416(1–2):33–46. doi:10.1007/s11010-016-2693-2
32. Ngan ES, Hashimoto Y, Ma ZQ, Tsai MJ, Tsai SY. Overexpression of Cdc25B, an androgen receptor coactivator, in prostate cancer. Oncogene. 2003;22(5):734–739. doi:10.1038/sj.onc.1206121
33. Lindqvist A, Kallstrom H, Karlsson Rosenthal C. Characterisation of Cdc25B localisation and nuclear export during the cell cycle and in response to stress. J Cell Sci. 2004;117(Pt 21):4979–4990. doi:10.1242/jcs.01395
34. Zhong Y, Yang J, Xu WW, et al. KCTD12 promotes tumorigenesis by facilitating CDC25B/CDK1/Aurora A-dependent G2/M transition. Oncogene. 2017;36(44):6177–6189. doi:10.1038/onc.2017.287
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.