Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
CircHIPK3 Alleviates High Glucose Toxicity to Human Renal Tubular Epithelial HK-2 Cells Through Regulation of miR-326/miR-487a-3p/SIRT1
Authors Zhuang L, Wang Z, Hu X, Yang Q, Pei X, Jin G
Received 30 October 2020
Accepted for publication 7 January 2021
Published 17 February 2021 Volume 2021:14 Pages 729—740
DOI https://doi.org/10.2147/DMSO.S289624
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ming-Hui Zou
Langen Zhuang, Ziwei Wang, Xiaolei Hu, Qingqing Yang, Xiaoyan Pei, Guoxi Jin
Department of Endocrinology, The First Affiliated Hospital of Bengbu Medical College, Bengbu, 233004 Anhui, People’s Republic of China
Correspondence: Guoxi Jin
Department of Endocrinology, The First Affiliated Hospital of Bengbu Medical College, Bengbu, Anhui, 233004, People’s Republic of China
Tel +86-18096530238
Email [email protected]
Background: The intervention of circular RNA HIPK3 (circHIPK3) in diabetes has drawn increasing attention in recent years. However, the underlying mechanism of circHIPK3 in diabetic nephropathy (DN) has not been fully elucidated. Thus, the current study aims to investigate the role of circHIPK3 in high glucose (HG)-induced toxicity to human renal tubular epithelial HK-2 cells.
Methods: The expression of circHIPK3 in HK-2 cells induced by HG was determined by qRT-PCR and Western blot. The regulatory effects of circHIPK3 and miR-326/miR-487a-3p on cells proliferative and apoptosis were evaluated by CCK-8 and flow cytometry. Dual-luciferase reporter assay was applied to predict the target genes of miR-326 or miR-487a-3p.
Results: Expression level of circHIPK3 in HK-2 cells was remarkably decreased after the treatment of HG. The overexpression of circHIPK3 effectively reversed the HG-induced HK-2 cell proliferation inhibition and apoptosis. Furthermore, SIRT1 was confirmed to be the target gene of miR-326 and miR-487a-3p, which were showed to be the downstream genes of circHIPK3. The silencing of miR-326 or miR-487a-3p was also proved to induce proliferation and reduce apoptosis in HG-induced HK-2 cells.
Conclusion: Our data suggest that overexpression of circHIPK3 can attenuate the proliferation inhibition of HK-2 induced by HG and inhibit apoptosis through sponging miR-326 or miR-487a-3p to regulate SIRT1.
Keywords: diabetic nephropathy, high glucose, circHIPK3, miR-326, miR-487a-3p, SIRT1
Introduction
Diabetic nephropathy (DN) is a devastating complication in patients with both type 1 and 2 diabetes mellitus (T1D and T2D) and has been the most common cause of end-stage renal disease all around the world.1,2 An increasing body of evidences point toward a mass of risk factors contributing to the development and progression of DN, including glucose metabolism disorder, long duration of diabetes, high blood pressure, obesity, and dyslipidemia3,4 .5 However, its specific molecular mechanism remains unclear, and there is still a demand for effective therapies.
Recently, circular RNAs (circRNA) have been proposed to be widely involved in the development and progression of human diseases.6,7 CircRNAs are types of newly identified noncoding RNAs (ncRNAs) with a stable closed-loop structure which are ubiquitous in mammals. CircHIPK3 is derived from HIPK3 (Homeodomain-interacting protein kinase 3) gene’s Exon2, which was identified as a tumor suppressor by reversing tumor-suppressive miRNAs and their target oncogenes in ovarian cancer.8 Inversely, it has been verified that the overexpression of circHIPK3 promotes the proliferative and invasive potentials of prostate cancer cells.9 Similarly, the downregulation of circHIPK3 was found to mediate HG-induced endothelial cell injury, and the significant upregulation of circHIPK3 regulated the endothelial proliferation and vascular dysfunction in DN.10 The specific role of circHIPK3 in DN, however, has not been clarified.
The most established function of circRNAs is that circRNAs have many miRNA-binding sites which can help them act as a sponge to miRNAs, leading the regulation of miRNAs on target genes, thereby altering the expression of these genes.11 MiRNAs are single-stranded RNAs, which are generated from pre-miRNAs by RNA polymerase II. The available evidence indicated that miRNAs are involved in the pathogenesis of DN and interacted with kidney cells.12 For instance, miRNAs play an important role in inflammation, apoptosis, autophagy, cell proliferation, and renal fibrosis in DN.13 MicroRNA crosstalk influences epithelial-to-mesenchymal, endothelial-to-mesenchymal, and macrophage-to-mesenchymal transitions in the kidney.14 MiRNA can suppress inflammation injury in HK-2 cells and urine-derived sepsis-induced kidney injury.15 MiR-326 was reported to play an important role in the progression of renal pathology, and miR-487a-3p functioned as a new tumor suppressor in prostate cancer, but its role in the progression of DN is still unclear.16,17 More importantly, miRNAs are also master regulators of EMT and dynamically regulate the balance between EMT and the reverse process, MET.18 Mechanically, kidney cells exploit miRNAs to regulate the EMT/MET by targeting different genes involved in EMT/MET process.19,20
Sirtuin 1 (SIRT1), a nicotinamide adenine dinucleotide (NAD+)–dependent histone deacetylase, plays a crucial role in the pathogenesis and development of DN.1 Accumulating evidences have demonstrated that the overexpression of SIRT1 could attenuate the development and progression of DN in different experiment models of diabetes and in renal cells, including podocytes, mesangial cells, and renal proximal tubular cells, incubated with high concentrations of glucose or advanced glycation end products (Role of sirtuin-1 in diabetic nephropathy).21 Mechanistically, SIRT1 plays a renoprotective role by exerting antioxidation and resisting apoptosis and inflammation.9 Additionally, the progression of DN was associated with the expression of miRNA, which targeted SIRT1 mRNA.
Collectively, the present study aims to investigate the role of circHIPK3 in high glucose-induced human renal tubular epithelial HK-2 cells, and miR-326/miR-487a-3p/SIRT1 pathway was studied for the further mechanisms.
Materials and Methods
Chemicals and Reagents
All antibodies were obtained from Cell Signaling Technology (Shanghai, China). Cell culture reagents were purchased from Gibco and Thermo Fisher Scientific (Switzerland). Puromycin, polybrene and Cell Counting Kit-8 (CCK-8) kit were provided by (CA1210, Solarbio, China).
Cell Culture and Treatment
Human renal tubular epithelial cell line HK-2 (ATCC® CRL-2190™) was purchased from American Type Culture Collection (ATCC). Cells were cultured in a humidified, 37°C, 5% CO2 incubator in Keratinocyte Serum Free Medium (K-SFM, Invitrogen, Carlsbad, CA, USA), which was supplied with 0.05 mg/mL bovine pituitary extract (BPE) and 5 ng/mL epidermal growth factor (EGF, Invitrogen). D-glucose was purchased from Sigma-Aldrich (St. Louis, MO, USA). For glucose treatment, HK-2 cells were treated by 45 mM D-glucose as HG treatment or 2 mM D-glucose as normal control for 48 h.
Cell Transfection
Transfection was performed when cell density reached 70%-80% by using Lipofectamine 3000 (Invitrogen, CA, USA) following the manufacturer’s instructions. MiR-326 mimics and inhibitor, miR-487a-3p mimics and inhibitor, negative control (miR-NC and NC-inhibitor), pcDNA-circHIPK3 overexpression vector (pcDNA-circHIPK3) and pcDNA negative control (pcDNA-NC), and SIRT1 siRNA (si-SIRT1) were synthesized by Genepharma Company (Shanghai, China). After transfection for 48 h, HK-2 cells were used for further assay.
Cell Viability Assay
The proliferation of HK-2 cells after treatment or transfection was detected by CCK-8 kit. HK-2 cells were seeded into 96-well plate with 5x103 cell/well in 10 μL CCK-8 kit reagent according to the manufacturer’s instructions. Two hours later, the optical density of each well at 450 nm wavelength (OD450) was examined by utilizing a microplate reader (Multiscan EX; Labsystems, Helsinki, Finland).
Cell Apoptosis Assay
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Beyotime Biotech, Haimen, China) was applied to assess apoptotic cells following the manufacturer’s instructions. HK-2 cells were washed and re-suspended in Binding Buffer, and then gently mixed with 10 μL FITC-AnnexinV and PI. After reaction, the ratio of apoptotic cells was assessed by flow cytometry within 1 h.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was conducted to measure the expression level of circHIPK3, miR-326, miR-487a-3p, and SITR1. TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was utilized to extract total RNA in serum of 60 donors and HK-2 cells, which treated with HG or transfection. Then, complementary DNAs (cDNAs) were generated by reverse transcription using Reverse Transcription kit (Takara, Tokyo, Japan). Afterwards, DNA amplification was conducted with SYBRSYBR® Premix Ex TaqTM (Takara) on the ABI 7500HT (Applied Biosystems). GAPDH was used as endogenous control, and the relative expression was measured using 2−ΔΔCt method. The primer sequences were listed as follows: circHIPK3, F 5’-TGGAGACTGGGGGAAGATGA-3’ and R 5’-CACACTAACTGGCTGAGGGG-3’; miR-326, F 5’-GCCTCTGGGCCCTTCCTC-3’ and R 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCT -GGAG-3’; miR-487a-3p, F 5’-ATGGCGGAATCATACAGGGAC-3’ and R 5’-CTCAACTGGTGTCGTGGAGTC-3’; SITR1, F 5’-AACAATTCCTCCACCTGAGC-3’ and R 5’-TCCCACAGGAGACAG AAACC-3’.
Western Blot Assay
The protein we used was extracted by utilizing RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA, USA). And Bicinchoninic acid (BCA) Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) was used for assessing the content of the proteins. Loaded equal amounts of proteins on SDS-GAGE and then transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Next, the membranes were blocked with 5% non-fat milk for 1 h, and incubated with applied primary antibody (dilution, 1:1000) overnight at 4°C, after that, using HRP-conjugated secondary antibodies (dilution, 1:1000) to incubate for 1 h. Subsequently, the membranes were visualized by using ECL kit (Sigma). After all, corresponding internal parameters were analyzed by Image LabTM Software (Bio-Rad Laboratories). Uncropped figures of the Western Blot Assay were shown in Supplementary figures S1-7.
Dual-Luciferase Reporter Gene Assay
3’UTR sequence of SIRT1-Wt and SITR-Mut were transcripted, and then, cloned circHIPK3-Wt and circHIPK3-Mut into the vector pGL3 containing the luciferase reporter gene. In addition, HK-2 cells were co-transfected with miR-326 mimics or miR-487a-3p mimics or negative control for 48 h, respectively. Dual-Luciferase reporter assay system (Promega, Madison, WI, USA) was utilized to investigate the luciferase activity.
Biotinylated RNA Pull-Down Assay
The pull-down assay with biotinylated RNA was performed as described previously.22 Briefly, for cirCHIPK3 pulled-down miRNAs, the biotinylated-circHIPK3 probe was incubated with C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA) generating probe-coated beads. Then, the coated beads were incubated with sonicated HK-2 cells at 4°C overnight. The bound RNA complexes were eluted from beads and purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The abundance of transcripts (miR-326 and miR-487a-3p) was evaluated by qRT-PCR analysis.
Statistical Analysis
All methods were performed in accordance with the relevant guidelines and regulations. Student’s t -test between two groups, and one-way ANOVA between multiple groups were performed to determine the significance of results as mean ± SD. Statistical analyses were performed using Graphpad Prism statistical software (Version 6.0; La Jolla, CA, USA). P values less than 0.05 were considered significant.
Results
HG-Treated HK-2 Cells Proliferation Inhibition and Apoptosis
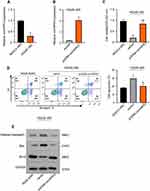
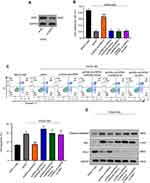
First, we established the HG-induced experiment models of human renal tubular epithelial HK-2 cells. HK-2 cells were cultured in medium with 5.5 mM glucose (normal glucose, “NG”) or 45 mM glucose (“HG”). CCK-8 assay showed that the cell ability was significantly inhibited after treatment with HG compared to NG (**p < 0.01.) (Figure 1A). Further, the flow cytometry assay proved that HG significantly induced HK-2 cells apoptosis after 48 h treatment compared to NG (**p < 0.01) (Figure 1B). Additionally, the expression of apoptotic proteins, caspase3, Bax and Bcl-2 was evaluated as showed in Figure 1C. Compared with NG, HG unregulated the expression of caspase3 and Bax, but conversely, Bcl-2 was downregulated in HK-2 cells after 48 h treatment. Thus, these results indicated that HG-induced cytotoxicity in the HK-2 cells through proliferation inhibition and promoting apoptosis.
CircHIPK3 Overexpression Alleviates HG-Induced Toxicity HK-2 Cells
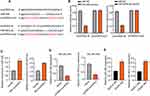
The expression of circHIPK3 on the serum of donors and HK-2 cells was detected by qRT-PCR. The results showed that circHIPK3 expression was down-regulated in the serum of DN patients compared with health control (**p < 0.01) (Figure 2A). Moreover, circHIPK3 expression was down-regulated with glucose concentration different (5.5 mM, 25 mM, 35 mM, 45 mM, 55 mM) after treatment for 48 h, the result indicated that circHIPK3 expression decreased gradually, reaching the lowest level at 45 mM, revealing that circHIPK3 might be associated with the damage of HK-2 cells (**p < 0.01) (Figure S1). In order to investigate the role of circHIPK3 on HG-induced HK-2 cells injury, HK-2 cells were transfected with pcDNA-negative control (vector) or pcDNA-circHIPK3 to elevate the level of circHIPK3. Expectedly, pcDNA-circHIPK3 transfection significantly up-regulated circHIPK3 expression in HK-2 cells (Figure 2B). Subsequently, CKK-8 assay indicated that pcDNA-circHIPK3 significantly promoted cell ability after HG treatment for 48 h in HK-2 cells compared to vector control (##p < 0.01). However, compared to NG, the cell ability was significantly inhibited in vector transfection after HG treatment (**p < 0.01) (Figure 2C).
Conversely, flow cytometry suggested that pcDNA-circHIPK3 significantly inhibited cell apoptosis compared to vector control after HG treatment for 48 h in HK-2 cells. However, compared with NG, the cell apoptosis was significantly promoted in vector transfection after HG treatment (**p < 0.01, #p < 0.05) (Figure 2D). Also, Western blot analysis implied that circHIPK3 overexpression up-regulated Bcl-2 expression but down-regulated caspase3 and Bax expression (Figure 2E), further indicating that circHIPK3 overexpression inhibited the apoptosis of HK-2 cells. Therefore, we demonstrated that circHIPK3 overexpression alleviated HG-induced cell injury by promoting HK-2 cells proliferation and inhibiting HK-2 cells apoptosis.
miR-326 and miR-487a-3p are Targets of circHIPK3
The molecular mechanism underlying the action of circHIPK3 on HK-2 cells was investigated in the further study. MiR-326 and miR-487a-3p were found to have the potential binding sites of circHIPK3 through the StarBase system (Figure 3A). Subsequently, a reduction of luciferase activity in HK-2 cells co-transfected with WT-circHIPK3 and miR-326 or miR-487a-3p was detected (**p < 0.01) (Figure 3B). To further determine this specific interaction, we performed a biotinylated RNA pull-down assay by using biotinylated circHIPK3 with biotinylated single-stranded antisense RNA as the control. The results exhibited that the enrichment of miR-326 and miR-487a-3p was increased by Bio-circHIPK3 compared with Bio-NC group (**p < 0.01) (Figure 3C). These results confirmed the direct interaction between circHIPK3 and miR-326 or miR-487a-3p. Also, we found that overexpression of circHIPK3 down-regulated the expression of miR-326 and miR-487a-3p in normal HK-2 cells as well as after HG treatment for 48 h (**p < 0.01) (Figure 3D, Figure S2). Reversely, the expression of miR-326 and miR-487a-3p was found up-regulated after treatment with HG in HK-2 cells (**p < 0.01) (Figure 3E). Thus, we verified that circHIPK3 was directly bound to miR-326 and miR-487a-3p, and negatively regulated its expression.
miR-326 or miR-487a-3p Alleviates HG-Induced Toxicity to HK-2 Cells
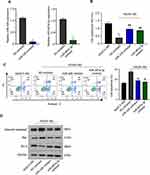
The function of miR-326 and miR-487a-3p in HG-induced cytotoxicity in HK-2 cells was further explored. HK-2 cells were transfected with NC-inhibitor, miR-326-inhibitor or miR-487a-3q-inhibitor, and a significant down-regulation of miR-326 or miR-487a-3q expression was observed in HK-2 cells after transfection with miR-326-inhibitor or miR-487a-3q-inhibitor, respectively (**p < 0.01) (Figure 4A). Afterwards, we found that NC-inhibitor suppressed the cell proliferation after HG treatment, but miR-326-inhibitor or miR-487a-3p-inhibitor induced the proliferation after HG treatment (**p < 0.01; ##p < 0.01) (Figure 4B). Also, miR-326-inhibitor or miR-487a-3p-inhibitor was found to suppress the HK-2 cells apoptosis after HG treatment (**p < 0.01) (Figure 4C). Moreover, the level of caspase3 and Bax was inhibited by miR-326-inhibitor or miR-487a-3p-inhibitor, but Bcl-2 was up-regulated in HG-induced HK-2 cells (p < 0.01) (Figure 4D). Together, miR-326 or miR-487a-3p down-regulation protected HK-2 cells against cytotoxicity of HG. Therefore, down-regulation miR-326 or miR-487a-3p alleviated HG-induced toxicity in HK-2 cells.
SIRT1 is a Target of miR-326 and miR-487a-3p in HK-2 Cells
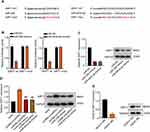
The molecular mechanism underlying the role of miR-326 and miR-487a-3p on HK-2 cells was investigated in the further study. SIRT1 was found to have the potential binding sites of miR-326 or miR-487a-3p through the StarBase system (Figure 5A). Subsequently, a reduction of luciferase activity in HK-2 cells overexpressed miR-326 or miR-487a-3p was detected (**p < 0.01) (Figure 5B). However, we found that its inhibition was disappeared after mutation of biding sites of SIRT1 (Figure 5B). To further determine this specific interaction, HK-2 cells were transfected with miR-326-mimics or miR-487a-3-mimics. The results showed that the mRNA and protein expression of SITR1 were seriously downregulated by miR-326-mimics or miR-487a-3-mimics, respectively (**p < 0.01) (Figure 5C). However, the upregulation of SITR1 was found in HK-2 cells after transfection with circHIPK3. Consistent with our results, SIRT1 upregulation has already been demonstrated in many diseases.23–25 Subsequently, co-transfection of circHIPK3 with miR-326-mimics or miR-487a-3-mimics led to the downregulation of mRNA and protein expression of SITR1 (**p < 0.01, ##p < 0.01) (Figure 5D). Moreover, the downregulation of SITR1 was also found in HK-2 cells after HG treatment compared to control (**p < 0.01) (Figure 5E). And for more accuracy, the mRNA and protein expression of SITR1 were detected in HK-2 cells transfection with circHIPK3 after HG treatment, and the downregulation of SITR1 was also found (**p < 0.01, Figure S3 and S4).
CircHIPK3 Regulates SIRT1 by Interacting with miR-326/miR-487a-3p in HG-Induced HK-2 Cells
Given that SITR1 was a target of miR-326 and miR-487a-3p, we explored the role of SIRT1 in HG-induced HK-2 cells. Firstly, down-regulated expression of SIRT1 protein was observed in HK-2 cells by siRNA SIRT1 (Figure 6A). Subsequently, CCK-8 assays showed that the cell ability was decreased via vector transfection after HG treatment compared to control, but the cell ability was promoted by transfecting with circHIPK3. Moreover, compared with circHIPK3 transfection after HG treatment, circHIPK3 co-transfection with miR-326 mimics or miR-487a-3p mimics and circHIPK3 transfection cooperated with silencing SIRT1 significantly inhibited the cell abilities, respectively (**p < 0.01, ##p < 0.01, ^^p < 0.01) (Figure 6B). Similarly, flow cytometry assays revealed that vector transfection after HG treatment induced HK-2 cells apoptosis, while circHIPK3 transfection reduced cell apoptosis. However, apoptosis was aggravated after circHIPK3 co-transfected with miR-326 mimics or miR-487a-3p mimics and circHIPK3 transfection cooperated with silencing SIRT1 (**p < 0.01, ##p < 0.01, ^^p < 0.01) (Figure 6C).
Furthermore, we also detected the protein expression related to apoptosis, including caspase3, Bax, and Bcl-2. Compared to control, caspase3 and Bax were up-regulated and Bcl-2 was down-regulated in HK-2 cells after HG treatment with vector transfection, by contrast, circHIPK3 transfection leading caspase3 and Bax down-regulation and Bcl-2 up-regulation; while compared to circHIPK3 transfection, co-transfection with miR-326 mimics or miR-487a-3p mimics and silencing SIRT1 up-regulated caspase3 and Bax but down-regulated Bcl-2 (p < 0.01) (Figure 6D). These data suggested that circHIPK3 regulates SIRT1 by interacting with miR-326/miR-487a-3p in HG-induced HK-2 cells.
Discussion
CircRNAs are a new class of noncoding RNAs and exert their functions via absorbing miRNA as miRNA sponges. In this study, we investigated the role of circHIPK3 in human renal tubular epithelial cells. CircHIPK3 could protect the HG-induced human renal tubular epithelial cells against toxicity. Mechanistically, circHIPK3 acts as an endogenous miR-326/miR-487a-3p sponge to regulate SIRT1 expression, thereby leading to reverse the high glucose-induced HK-2 cell proliferation inhibition and apoptosis.
DN is a chronic complication of diabetes mellitus, which is typically characterized by renal tubular damage in patients.26,27 In our study, renal tubular epithelial HK-2 cells were treated by HG to cause renal tubular epithelial cells damage. Thus, we found HG decreased HK-2 cells ability, while induced apoptosis validated by flow cytometry and Western blot analysis. These results were in line with previous study that HG treatment induced HK-2 apoptosis.28 Therefore, the HG-induced HK-2 cell was a nice model to mimic epithelial cells damage of DN in further study.
Previous studies showed that circHIPK3 mediated retinal vascular dysfunction in diabetes mellitus.29 In our data, we found the level of circHIPK3 in serum of DN patients was downregulated compared with health control, which was keeping in line with the study of group.9,11 In HK-2 cells, circHIPK3 was down-regulated after HG treatment, suggesting that HG-treated cells presented similar feature with DN patients. Moreover, Cao et al showed that HG could also induce circHIPK3 downregulation in human umbilical vein endothelial cells and in primary aortic endothelial cells from diabetic patients.10 The downregulation of circHIPK3 mediated HG-induced vascular endothelial cell injury plays a pivotal role in initiation and progression of cardiovascular complications of diabetes mellitus.10,30,31
Overexpression of circHIPK3 markedly promoted proliferation, but suppressed apoptosis of HK-2 cells even stressed by HG. Thus, overexpression of circHIPK3 could reverse the cell damage caused by HG. Previous reports revealed that circHIPK3 plays a vital role in various cancers.5,21 Consistent with our experimental results, studies have shown that the expression of Bcl-2 was up-regulated and Bax was down-regulated by circHIPK3.32,33 Moreover, circHIPK3 could function through sponging miRNAs in the progression of many cancers.34,35 Thus, we speculated that circHIPK3 served as a competing endogenous RNAs to regulate its target miRNA, thus affecting the cell ability of HK-2 cells.
In this study, luciferase reporter assays showed that the high level of circHIPK3 was associated with the decreased expression of miR-326 and miR-487a-3p, and decreased circHIPK3 level was related to increased expression of miR-326 and miR-487a-3p, suggesting that circHIPK3 was targeting miR-326 and miR-487a-3p. Moreover, HG was also found to induce miR-326 and miR-487a-3p expression, which was consistent with the expression of circHIPK3 under HG treatment. Interestingly, the decreased cell ability caused by HG was slowed under treatment with miR-326 or miR-487a-3p inhibitor; meanwhile, the apoptosis caused by HG was decreased by treatment with miR-326 or miR-487a-3p inhibitor.
It was reported that miR-326 may be correlated with obstructive renal injury, and be regarded as potential biomarkers for disease diagnosis or prognosis monitoring,36 as well as miR-487a-3p. However, the role of miR-326 and miR-487a-3p mediated by circHIPK3 in DN was still unknown.
Furthermore, the expression of SIRT1, the target gene of miR-326/miR-487a-3p, was upregulated after circHIPK3 overexpression in HK-2 cells. Further experiments confirmed that overexpression of miR-326 or miR-487a-3p could down-regulate the level of SIRT1, thus abolishing the influence of circHIPK3 overexpression in HK-2 cells. These results suggested that circHIPK3 regulated the expression of SIRT1 by interacting with miR-326 or miR-487a-3p, thereby influencing the proliferation and apoptosis of HG-induced HK-2 cells. Overexpression of circHIPK3 promotes the proliferation and suppresses apoptosis of HG-induced HK-2 cells, which revealed that overexpression of circHIPK3 alleviates high glucose toxicity to human renal tubular epithelial cells through sponging miR-326/miR-487a-3p to regulate SIRT1 expression. Our results also indicated that circHIPK3 acts as a potential broad-spectrum suppressive gene and deserves more attention on its biological and clinical relevance in various types of diseases. Further characterization of factors influencing circHIPK3 expression or its binding may be introduced to yield new treatment strategies for diseases, especially for patients with DN.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Lv L, Zhang J, Tian F, Li X, Li D, Yu X. Arbutin protects HK-2 cells against high glucose-induced apoptosis and autophagy by up-regulating microRNA-27a. Artif Cells Nanomed Biotechnol. 2019;47(1):2940–2947. doi:10.1080/21691401.2019.1640231
2. Lai J, Xin J, Fu C, Zhang W. CircHIPK3 promotes proliferation and metastasis and inhibits apoptosis of renal cancer cells by inhibiting MiR-485-3p. Cancer Cell Int. 2020;20(1):248. doi:10.1186/s12935-020-01319-3
3. Tang X, Sun G, He Q, et al. Circular noncoding RNA circMBOAT2 is a novel tumor marker and regulates proliferation/migration by sponging miR-519d-3p in colorectal cancer. Cell Death Dis. 2020;11(8):625. doi:10.1038/s41419-020-02869-0
4. Diekmann U, Naujok O. Circular non-coding RNAs in diabetic retinopathy. Non Coding RNA Invest. 2017;1:16. doi:10.21037/ncri.2017.11.01
5. Teng F, Xu J, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;20:8–17. doi:10.3390/ijms20133153
6. Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab. 2019;47:69–83. doi:10.1080/21691401.2019.1616552
7. Jin J, Sun H, Shi C, et al. Circular RNA in renal diseases. J Cell Mol Med. 2020;24(12):6523–6533. doi:10.1111/jcmm.15295
8. Xie Y, Yuan X, Zhou W, et al. The circular RNA HIPK3 (circHIPK3) and its regulation in cancer progression: review. Life Sci. 2020;254:117252. doi:10.1016/j.lfs.2019.117252
9. Qi C, Mao X, Zhang Z, Wu WH. Classification and Differential Diagnosis of Diabetic Nephropathy. J Diabetes Res. 2000;7:8637138. doi:10.1038/sj.cdd.4400714
10. Cao Y, Yuan G, Zhang Y, Lu R. High glucose-induced circHIPK3 downregulation mediates endothelial cell injury. Biochem Biophys Res Commun. 2018;507(1–4):362–368. doi:10.1016/j.bbrc.2018.11.041
11. Cui G, Wang L, Huang W. Circular RNA HIPK3 regulates human lens epithelial cell dysfunction by targeting the miR-221-3p/PI3K/AKT pathway in age-related cataract. Exp Eye Res. 2020;198:108128. doi:10.1016/j.exer.2020.108128
12. Zurawek M, Dzikiewicz-Krawczyk A, Izykowska K, et al. miR-487a-3p upregulated in type 1 diabetes targets CTLA4 and FOXO3. Diabetes Res Clin Pract. 2018;142:146–153. doi:10.1016/j.diabres.2018.05.044
13. Lv J, Wu Y, Mai Y, Bu S. Noncoding RNAs in Diabetic Nephropathy: pathogenesis, Biomarkers, and Therapy. J Diabetes Res. 2020;2020:3960857. doi:10.1155/2020/3960857
14. Srivastava SP, Hedayat AF, Kanasaki K, Goodwin JE. microRNA Crosstalk Influences Epithelial-to-Mesenchymal, Endothelial-to-Mesenchymal, and Macrophage-to-Mesenchymal Transitions in the Kidney. Front Pharmacol. 2019;10:904. doi:10.3389/fphar.2019.00904
15. Shen J, Liu L, Zhang F, Gu J, Pan G. Lnc RNA Tap SAKI promotes inflammation injury in HK −2 cells and urine derived sepsis-induced kidney injury. J Pharm Pharmacol. 2019;71(5):839–848. doi:10.1111/jphp.13049
16. Xia Y, Tao JH, Fang X, et al. MicroRNA-326 Upregulates B Cell Activity and Autoantibody Production in Lupus Disease of MRL/LPR Mice. Mol Ther Nucleic Acids. 2018;11:284–291. doi:10.1016/j.omtn.2018.02.010
17. Wang M, Yu W, Gao J, et al. MicroRNA-487a-3p functions as a new tumor suppressor in prostate cancer by targeting CCND1. J Cell Physiol. 2020;235(2):1588–1600. doi:10.1002/jcp.29078
18. Ding XM. MicroRNAs: regulators of cancer metastasis and epithelial-mesenchymal transition (EMT). Chin J Cancer. 2014;33(3):140–147. doi:10.5732/cjc.013.10094
19. Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123(1):150–163. doi:10.1172/JCI64946
20. Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58(2):629–641. doi:10.1002/hep.26369
21. Ni H, Li W, Zhuge Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. 2019;292:188–196. doi:10.1016/j.ijcard.2019.04.006
22. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-0454-8
23. Alves-Fernandes DK, Jasiulionis MG. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int J Mol Sci. 2019;20(13):20. doi:10.3390/ijms20133153
24. Ji K, Sun X, Liu Y, et al. Regulation of Apoptosis and Radiation Sensitization in Lung Cancer Cells via the Sirt1/NF-kappaB/Smac Pathway. Cell Physiol Biochem. 2018;48(1):304–316. doi:10.1159/000491730
25. Chen J, Cao L, Li Z, Li Y. SIRT1 promotes GLUT1 expression and bladder cancer progression via regulation of glucose uptake. Hum Cell. 2019;32(2):193–201. doi:10.1007/s13577-019-00237-5
26. Lv L, Li D, Tian F, Li X, Jing Z, Yu X. Silence of lncRNA GAS5 alleviates high glucose toxicity to human renal tubular epithelial HK-2 cells through regulation of miR-27a. Artif Cells Nanomed Biotechnol. 2019;47(1):2205–2212. doi:10.1080/21691401.2019.1616552
27. Flyvbjerg A. The role of the complement system in diabetic nephropathy. Nat Rev Nephrol. 2017;13(5):311–318. doi:10.1038/nrneph.2017.31
28. Zhang J, Jiang T, Liang X, et al. lncRNA MALAT1 mediated high glucose-induced HK-2 cell epithelial-to-mesenchymal transition and injury. J Physiol Biochem. 2019;75(4):443–452. doi:10.1007/s13105-019-00688-2
29. Shan K, Liu C, Liu B-H, et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation. 2017;136:1629–1642. doi:10.1161/circulationaha.117.029004
30. Rahman S, Rahman T, Ismail AA, Rashid AR. Diabetes-associated macrovasculopathy: pathophysiology and pathogenesis. Diabetes Obes Metab. 2007;9(6):767–780. doi:10.1111/j.1463-1326.2006.00655.x
31. Vinik A, Flemmer M. Diabetes and macrovascular disease. J Diabetes Complications. 2002;16(3):235–245. doi:10.1016/s1056-8727(01)00212-4
32. Zhang Y, Li C, Liu X, et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288. doi:10.1016/j.ebiom.2019.09.051
33. Gaumer S, Guénal I, Brun S, Théodore L, Mignotte B. Bcl-2 and Bax mammalian regulators of apoptosis are functional in Drosophila. Cell Death Differ. 2000;7(9):804–814. doi:10.1038/sj.cdd.4400714
34. Ghasemi S. Down-regulation of circular RNAITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. 2019;49(2):687–695. doi:10.3906/sag-1806-50
35. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi:10.1016/j.bbrc.2018.10.087
36. Wang S, Wu L, Du L, Lu H, Chen B, Bai Y. Reduction in miRNA-125b-5p levels is associated with obstructive renal injury. Biomed Rep. 2017;6(4):449–454. doi:10.3892/br.2017.875
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.