Back to Journals » Cancer Management and Research » Volume 13
CircGDI2 Regulates the Proliferation, Migration, Invasion and Apoptosis of OSCC via miR-454-3p/FOXF2 Axis
Authors Shi D, Li H, Zhang J, Li Y
Received 14 August 2020
Accepted for publication 10 December 2020
Published 11 February 2021 Volume 2021:13 Pages 1371—1382
DOI https://doi.org/10.2147/CMAR.S277096
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Dan Shi,1 Huiyun Li,2 Junge Zhang,3 Yadong Li1
1Department of Oral Medicine Centre, Henan Provincial People’s Hospital, Zhengzhou, Henan, People’s Republic of China; 2Department of Anesthesiology, Henan Provincial People’s Hospital, Zhengzhou, Henan, People’s Republic of China; 3Department of Ophthalmology, Henan Provincial People’s Hospital, Zhengzhou, Henan, People’s Republic of China
Correspondence: Yadong Li
Department of Oral Medicine Centre, Henan Provincial People’s Hospital, No. 7 Wei-Wu Road, Zhengzhou City, 450000 Henan Province, People’s Republic of China
Tel +86-186-38723618
Email [email protected]
Background: Aberrant expression of circular RNA (circRNA) is involved in the occurrence and development of multifarious cancers, including oral squamous cell carcinoma (OSCC). However, the biological role of circGDI2 and the action mechanism in OSCC remain largely unclear.
Methods: The expression levels of circGDI2, miR-454-3p and forkhead box F2 (FOXF2) were examined by quantitative real-time PCR (qRT-PCR) or Western blot. The stability of circGDI2 was confirmed by Ribonuclease R (RNase R) assay. Cell Counting Kit 8 (CCK8) assay, colony formation and transwell assay were used to detect cell proliferation, migration or invasion. Cell apoptosis was tested by flow cytometry. Dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were employed to verify the interaction between miR-454-3p and circGDI2 or FOXF2. Moreover, xenograft mouse models were constructed to assess tumor growth in vivo.
Results: CircGDI2 was a stable circRNA and was low expressed in OSCC tissues and cells. CircGDI2 overexpression could effectively inhibit the proliferation, migration, invasion and promote apoptosis in OSCC cells, and suppress OSCC tumor growth in nude mice in vivo. MiR-454-3p could be sponged by circGDI2, and its overexpression could mitigate the suppressive effects of circGDI2 overexpression on OSCC progression. In addition, FOXF2 was a target of miR-454-3p, and miR-454-3p silence could impede the cell growth of OSCC cells by enhancing FOXF2 expression. Meanwhile, circGDI2 positively regulated FOXF2 expression by targeting miR-454-3p.
Conclusion: CircGDI2 served as a repressor to restrain OSCC malignancy via miR-454-3p/FOXF2 axis, which might be a novel biomarker for targeted OSCC therapy.
Keywords: OSCC, circGDI2, miR-454-3p, FOXF2
Introduction
Oral squamous cell carcinoma (OSCC) is an aggressive and metastatic malignant oral malignancy with rising incidence.1,2 Nowadays, although much progress has been made in diagnosis and management, but ascribe to the lack of early diagnostic markers, later diagnosis at an advanced stage and high recurrence rate, the survival rate of OSCC patients is still disappointing.3 As a gene-related disease, the precise molecular mechanism and hereditary basis under OSCC tumorigenesis are still quite enigmatic. Therefore, it is of crucial importance to find diagnostic strategies and identify therapeutic targets to improve OSCC treatment.
Circular RNAs (circRNAs) are a group of regulatory RNAs with low or no protein-coding potentiality and broadly present in eukaryotes.4 Recently, circRNAs have gradually become a hot research topic in the field of disease research. Numerous studies indicate that the unbalance of circRNAs is concerned with the carcinogenesis of multifarious cancers, including OSCC.5,6 For example, circ_0008309 and circ-PKD2 were downregulated in OSCC tissues and could impede OSCC carcinogenesis.7,8 Similarly, circGDI2, also known as circ_0005379, was distinctly downregulated in OSCC tissues and cells and hindered OSCC cell proliferation and metastasis.9 However, the function and regulatory mechanism of circGDI2 in OSCC malignant behaviors remain largely unclear.
MicroRNAs (miRNAs) are one type of small noncoding RNAs, which are highly conserved across species and function in posttranscriptional regulation of gene expression.10 MiRNAs are pivotal regulators of many cancers and could serve as potential biomarkers for diagnosis and therapy, including breast cancer,11 prostate cancer12 and pancreatic cancer.13 Interestingly, miR-454-3p acts as a tumor depressor or promoter in different cancers.14,15 Guo et al certified that miR-454 was upregulated and facilitated OSCC cells growth by sponging NR3C2.16 The transcription regulator forkhead box F2 (FOXF2) is a critical molecule that participates in embryogenesis and tumorigenic processes.17 FOXF2 level was lessened in OSCC tissues and FOXF2 silence signally accelerated OSCC cells progression.18 However, whether FOXF2 was implicated in miR-454-3p-mediated OSCC progression has not been reported.
In this study, the expression patterns of circGDI2, miR-454-3p and FOXF2 were first analyzed in OSCC tissues and cells. Then, the functional complementation analysis was conducted to verify the functions of them and the relationships among the circRNA/miRNA/mRNA networks. This study might provide a novel basis and biomarker for OSCC treatment.
Materials and Methods
Clinical Samples
Thirty-four pairs of tumor tissues and adjacent non-tumor tissues were procured from OSCC patients who underwent surgery and enrolled at Henan Provincial People’s Hospital. No patients received treatments before surgery resection. Before conducting this project, all participates were completely informed about this research purpose and signed informed consents. This study was empowered by the Institutional Ethics Committee of Henan Provincial People’s Hospital and implemented keeping to the Declaration of Helsinki.
Cell Culture
Human OSCC cell lines (SCC-15 and HSC-3) cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) and normal oral keratinocyte (HOK) cells cultured in oral keratinocyte growth medium (ScienCell, Carlsbad, CA, USA) were all purchased from Bena Culture Collection (Beijing, China). All the media were supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% antibiotics (100 units/mL; Invitrogen, Camarillo, CA, USA). Cells were maintained in an environment filled with 5% CO2 at 37°C.
Quantitative Real-Time PCR (qRT-PCR)
TRIzol reagent (Invitrogen) was applied for isolation of RNA from OSCC tissues and cells. The cDNA was synthesized using PrimeScript RT Reagent Kit (for circGDI2, GDP dissociation inhibitor (GDI2) and FOXF2; Takara, Dalian, China) and Bulge-Loop miRNA qRT-PCR Starter Kit (for miR-454-3p; RiboBio, Guangzhou, China). Then, qRT-PCR was performed on a PCR system using Fast SYBR Green Master Mix (Applied Biosystems, Austin, TX, USA). The relative enrichments of circGDI2, GDI2, miR-454-3p and FOXF2 were calculated by 2−ΔΔCt method after normalization with reference control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6. The special primer sequences (5ʹ-3ʹ) synthesized from Sangon Biotech (Shanghai, China) were displayed as follows: circGDI2, forward: GCCCATACCTTTATCCACTC and reverse: GTCAACATTCCAGTCTCTTCCT; GDI2, forward: ATTCCACAGAACCAAGTCAATCGA and reverse: CTTCTCAGGCTCCTTGGTTTCC; miR-454-3p, forward: GCGCGTAGTGCAATATTGCTTA and reverse: AGTGCAGGGTCCGAGGTATT; FOXF2 forward: AATGCCACTCGCCCTACAC and reverse: CGTTCTGGTGCAAGTAGCTCT; GAPDH, forward: GCACCGTCAAGGCTGAGAAC and reverse: TGGTGAAGACGCCAGTGGA; U6, forward: GCTTCGGCAGCACATATACTAAAAT, and reverse: CGCTTCACGAATTTGCGTGTCAT.
Ribonuclease R (RNase R) Assay
For RNase R test, the RNA was incubated at 37°C with or without RNase R (Epicentre Technologies, Madison, WI, USA) for 15 min. The negative control without RNase R addition was named as Mock. Then, the RNA expression of circGDI2 and linear GDI2 were determined by qRT-PCR.
Cell Transfection
For the overexpression of circGDI2 and miR-454-3p, pCD-ciR-circGDI2 overexpression vector (oe-circGDI2) or miR-454-3p mimic was transduced into SCC-15 and HSC-3 cells, with pCD-ciR empty vector (Vector) or miR-NC mimic as negative control, respectively. For silencing FOXF2 and miR-454-3p, small interfering RNA against FOXF2 (si-FOXF2) or miR-454-3p inhibitor (anti-miR-454-3p) was, respectively, introduced into SCC-15 and HSC-3 cells, with si-NC or anti-miR-NC as negative control. The above oligonucleotides (50 nM) or plasmids (4 μg) obtained from Geneseed (Guangzhou, China) or RiboBio were transduced into cells via Lipofectamine 3000 (Invitrogen) referring to specifications.
Cell Counting Kit 8 (CCK8) Assay
CCK8 assay was carried out to assess cell viability. In brief, SCC-15 or HSC-3 cells were harvested and plated into 96-well plates at 24 h post transfection. At the indicated times, 10 μL CCK8 reagent (Beyotime Biotechnology, Shanghai, China) was added. After incubation at 37°C for 4 h, the absorbance at 450 nm was determined by a microplate reader (Bio-Rad, Hercules, CA, USA).
Colony Formation Assay
After transfection, SCC-15 or HSC-3 cells (1000 cells/well) were plated into 6-well plates and routinely incubated for 2 weeks. Afterwards, the media were removed and the generated colonies (cell mass containing more than 50 cells) were dyed with 0.5% Crystal Violet Staining Solution (Beyotime Biotechnology) for 30 min after fixation. Finally, the colonies were photographed and counted using a microscope.
Flow Cytometry
For apoptosis detection, SCC-15 and HSC-3 cells after 48 h transfection were harvested and re-suspended in binding buffer (Beyotime Biotechnology), and then stained with 5 μL Annexin V-fluorescein isothiocyanate (Annexin V-FITC, BD Biosciences, San Jose, CA, USA) and 10 μL propidine iodide (PI; BD Biosciences) for 15 min in a dark place on the basis of the supplier’s instructions. Subsequently, the apoptotic cells (FITC+/PI±) were determined by a flow cytometer (BD Biosciences).
Transwell Migration and Invasion Assays
Transwell chambers (Corning Incorporate, Corning, NY, USA) with an 8-μm pore membrane enveloped with Matrigel (Corning Incorporate) or not were applied for invasion or migration analysis, respectively. The transfected SCC-15 and HSC-3 cells re-suspended in serum-free medium were seeded into the upper chambers. The lower chambers were filled with complete medium. Twenty-four hour later, the cells on the surface of the upper chambers were gently removed, and the metastatic cells through the membrane were dyed with Crystal Violet Staining Solution (Beyotime Biotechnology) after immobilization. The cell number of migration or invasion was counted under a light microscope (amplification: 100 ×).
Dual-Luciferase Reporter Assay
The wide-type (WT) sequence of circGDI2 or FOXF2 3ʹ-untranslated region (3ʹUTR) containing miR-454-3p binding sites and the corresponding mutant (MUT) sequence of circGDI2 or FOXF2 3ʹUTR harboring the mutant binding sites with miR-454-3p were inserted into pGL3 luciferase promoter plasmids (Promega, Madison, WI, USA), termed as circGDI2-WT, FOXF2 3ʹUTR-WT, circGDI2-MUT or FOXF2 3ʹUTR-MUT. Afterwards, SCC-15 and HSC-3 cells were introduced with generated luciferase reporter plasmids and miR-454-3p mimic or miR-NC mimic. After transfection for 48 h, a Dual-Luciferase Reporter Assay System (Promega) was adopted to evaluate the relative luciferase activity.
RNA Immunoprecipitation (RIP) Assay
SCC-15 and HSC-3 cells were lysed via RIP lysis buffer (Bio-Rad) at 48 h post transfection. Subsequently, the cell lysate was incubated overnight at 4°C with Protein A/G Magnetic beads (Pierce, Rockford, IL, USA) containing argonaute 2 antibody (anti-Ago2; Abcam, Cambridge, UK) or immunoglobulin G antibody (anti-IgG; Abcam). The RNA enrichments of circGDI2, miR-454-3p and FOXF2 from purified RNA immune-precipitate were tested by qRT-PCR.
Western Blot (WB) Analysis
After isolation of protein from tissues and cells by NP-40 Lysis Buffer (Beyotime Biotechnology), the BCA Protein Assay Kit (Beyotime Biotechnology) was utilized for protein quantification. Protein (30 μg) was isolated by 10% SDS-PAGE gel and then electrotransferred onto polyvinylidene fluoride (PVDF) membranes (Invitrogen). After sealing with 5% fat-free milk for 2 h, the membranes were added with the first antibody against FOXF2 (ab198283; Abcam; 1: 1000) or control GAPDH (ab181602; Abcam; 1: 10,000) overnight at 4°C and then added with horseradish peroxidase (HRP)-conjugated secondary antibody (ab205718; Abcam; 1: 5000) for 1 h. Last, protein signals were visualized by Enhanced Chemiluminescence Kit (Millipore, Billerica, MA, USA).
Xenograft Tumor Model
A total of 12 BALB/c nude mice (male, 5-weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). For in vivo assay, the mice were divided into oe-circGDI2 group (n=6) and Vector group (n=6) at random, and HSC-3 cells stably overexpressing oe-circGDI2 or Vector in PBS were subcutaneously inoculated into the left flank of mice for establishing xenograft model. The length (L) and width (W) of the generated tumors were measured by caliper every 5 days until the mice were sacrificed at day 30. The tumor volume (V) was computed by the formula of (length × width2)/2 and the weight of tumors was recorded at 30 d post inoculation. The enrichments of circGDI2, miR-454-3p and FOXF2 in the tumors were detected by qRT-PCR or Western blot analysis. All experiments performed on nude mice were authorized by the Institutional Animal Care and Use Committee (IACUC) of Henan Provincial People’s Hospital and implemented following the Management and Use Guidelines for Laboratory Animals of National Institutes of Health (NIH).
Statistical Analysis
All assays were conducted at least thrice, and all results are presented as the mean ± standard deviation and analyzed through GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, CA, USA). Differences between two groups or among multiple groups were compared via Student’s t-test or one-way analysis of variance followed by Tukey’s test. Pearson correlation analysis was performed to analyze the correlation among circGDI2, miR-454-3p, and FOXF2 in OSCC samples. P < 0.05 was defined to be significantly different.
Results
The Expression and Characterization of circGDI2 in OSCC Tumor Tissues and Cells
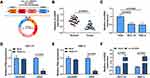
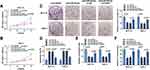
CircGDI2, also known as circ_0005379, is located on chr10:5,827,104–5,842,668 of chromosome and is derived from exons 2 to 5 of the GDI2 gene with the spliced mature sequence length of 674 bp (Figure 1A). Initially, this study investigated the expression of circGDI2 in OSCC tumor tissues and cells using qRT-PCR assay. The results suggested that circGDI2 was markedly low expressed in OSCC tumor tissues and two OSCC cell lines (SCC-15 and HSC-3) compared to adjacent normal tissues and HOK cells (Figure 1B and C). Through RNase R assay, it was verified that circGDI2 could resist RNase R digestion and was more stable than linear GDI2 mRNA (Figure 1D and E), implying that circGDI2 was indeed a circRNA.
Overexpression of circGDI2 Inhibited the Proliferation, Migration and Invasion in OSCC Cells
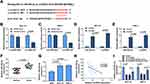
To illuminate the role of circGDI2 in OSCC progression, the gain-of-function experiments were performed using oe-circGDI2. After transfection of oe-circGDI2 into SCC-15 and HSC-3 cells, the high transfection efficiency of circGDI2 overexpression was confirmed through the raised expression of circGDI2 (Figure 1F). CCK8 and clone formation assay revealed that the proliferation of SCC-15 and HSC-3 cells was distinctly inhibited by circGDI2 overexpression, indicated by reduced cell vitality (Figure 2A and B) and colony number (Figure 2C). Meanwhile, the apoptosis of SCC-15 and HSC-3 cells was dramatically incremental (Figure 2D), and the numbers of migrated and invaded SCC-15 and HSC-3 cells were inhibited after circGDI2 introduction (Figure 2E and F).
CircGDI2 Served as a Molecular Sponge of miR-454-3p
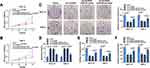
For exploring the potential molecular mechanisms of circGDI2 in OSCC tumorigenesis, starBase software was utilized to find the probable targets of circGDI2. As depicted in Figure 3A, the putative targeting sites between circGDI2 and miR-454-3p were exhibited. Then, circGDI2-WT/MUT reporter vectors were constructed based on the binding sequence of circGDI2 and miR-454-3p. Dual-luciferase reporter assay results manifested that miR-454-3p overexpression could distinctly hinder the luciferase activity of SCC-15 and HSC-3 cells transfected with circGDI2-WT, while it had no effect on that of the circGDI2-MUT group (Figure 3B and 3C). Meanwhile, RIP assay also demonstrated that the enrichments of circGDI2 and miR-454-3p were markedly enhanced in the anti-Ago2 group (Figure 3D and E), which further verified the combination between them. Contrary to circGDI2 expression, miR-454-3p level was found to be upregulated in OSCC tissues and cells (Figure 3F and G) and reversely correlated with that of circGDI2 in OSCC tissues (Figure 3H). Besides, circGDI2 overexpression had a strong inhibitory effect on miR-454-3p expression, while the decrease was signally alleviated by miR-454-3p addition (Figure 3I). These data demonstrated that circGDI2 could directly target miR-454-3p in OSCC cells.
CircGDI2 Could Inhibit OSCC Progression by Regulating miR-454-3p
The rescue experiments were carried out to confirm whether circGDI2 impeded OSCC progression by targeting miR-454-3p. The overexpression vectors of oe-circGDI2 and miR-454-3p mimic were co-transfected into SCC-15 and HSC-3 cells. The results proposed that the inhibitory effects of oe-circGDI2 on cell viability (Figure 4A and B), colony formation (Figure 4C), migration (Figure 4E), and invasion (Figure 4F) and the promotional effect on cell apoptosis (Figure 4D) were all partially alleviated by miR-454-3p overexpression. Therefore, these findings indicated that circGDI2 inhibited cell proliferation, migration, invasion and promoted cell apoptosis by sponging miR-454-3p.
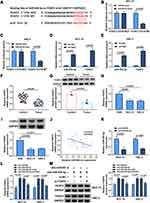
FOXF2 Was a Target of miR-454-3p
To identify the potential target gene of miR-454-3p in OSCC, bioinformatics software starBase was used. The software revealed that miR-454-3p contained the complementary sites with the 3ʹUTR of FOXF2 mRNA (Figure 5A). The interaction between miR-454-3p and FOXF2 was certified by dual-luciferase reporter assay and RIP assay. The results showed that the luciferase activity of FOXF2 3ʹUTR-WT reporter vector could be apparently reduced by miR-454-3p overexpression, while the decrease was abolished in that of FOXF2 3ʹUTR-MUT vector (Figure 5B and C). Meanwhile, the results of RIP assay suggested that the levels of miR-454-3p and FOXF2 were remarkably increased in the anti-Ago2 group (Figure 5D and E). In line with circGDI2, the mRNA and protein expression of FOXF2 was obviously downregulated in OSCC tumor tissues and cells (Figure 5F–I). Furthermore, the miR-454-3p level was negatively related to FOXF2 level in OSCC tumor tissues (Figure 5J). Then, the effect of miR-454-3p on FOXF2 expression was explored. The transfection efficiency of miR-454-3p inhibition in OSCC cells was first verified (Figure 5K). In addition, the expression of FOXF2 was facilitated by miR-454-3p inhibitor, and the effect was overturned by FOXF2 silence (Figure 5L and M). Taken together, this evidence strongly certified the direct targeted relationship between FOXF2 and miR-454-3p in OSCC cells.
Knockdown of FOXF2 Inverted the Hindered Effect of miR-454-3p Inhibitor in OSCC
To determine whether miR-454-3p could regulate OSCC progression by mediating FOXF2 expression, OSCC cells were transfected with anti-miR-454-3p, anti-miR-454-3p + si-FOXF2, or corresponding controls. The results of CCK8, clone formation and transwell assays suggested that the proliferation (Figure 6A–C), migration (Figure 6E) and invasion (Figure 6F) of SCC-15 and HSC-3 cells were visibly suppressed after miR-454-3p silence, while the effects were reversed by FOXF2 downregulation. Meanwhile, FOXF2 silencing also mitigated the enhancive effect of anti-miR-454-3p on cell apoptosis (Figure 6D). Altogether, these findings demonstrated that FOXF2 repressed the progression of OSCC, which was in keeping with the function of circGDI2 in OSCC.
CircGDI2 Could Regulate FOXF2 Expression by Sponging miR-454-3p
To confirm the correlation among circGDI2, miR-454-3p and FOXF2, we transfected oe-circGDI2, oe-circGDI2 and miR-454-3p mimic or corresponding controls into SCC-15 and HSC-3 cells. Through the measure of FOXF2 expression, it was discovered that circGDI2 overexpression dramatically enhanced the mRNA and protein levels of FOXF2, while this effect was visibly canceled by miR-454-3p addition (Figure 7A and B). Hence, these results revealed that circGDI2 could improve FOXF2 expression by inhibiting miR-454-3p.
Overexpression of circGDI2 Suppressed OSCC Tumor Growth in vivo
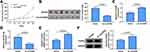
To further investigate the effect of circGDI2 in OSCC tumorigenesis in vivo, HSC-3 cells with stable circGDI2 overexpression were subcutaneously injected into nude mice to construct the mice xenograft models. Compared with those of control xenografts (Vector group), the tumor volume and weight of nude mice in oe-circGDI2 group were markedly reduced over a period of 30 days (Figure 8A and B). Meanwhile, the expression of circGDI2 was increased while miR-454-3p expression was notably declined in oe-circGDI2 group (Figure 8C and D). Congruously, the mRNA and protein expression of FOXF2 was clearly enhanced (Figure 8E and F) after circGDI2 overexpression. Collectively, the in vivo mice experiments suggested that circGDI2 inhibited tumor formation by regulating the miR-454-3p/FOXF2 axis.
Discussion
OSCC is a common malignant tumor influencing the head and neck region that damages oral epithelial cells.19 It is urgent to seek molecular biomarkers and therapeutic methods for OSCC diagnosis and remedy, thus to improve the survival and life quality of OSCC patients. In this present study, we found the dysregulated expression of circGDI2, miR-454-3p and FOXF2 in OSCC, implying their potential roles in OSCC development. Meanwhile, we established a novel regulatory network of the circGDI2/miR-454-3p/FOXF2 axis and explored its action mechanism in OSCC etiology.
CircRNAs could play an imperative regulatory role in OSCC progression through facilitating or inhibiting cancer development via sponging miRNAs.6 For instance, circRNA_100290 was upregulated in OSCC and could promote OSCC carcinogenesis via acting as a decoy of miR-29b,20 while circ-PKD2 was low expressed in OSCC and could inhibit OSCC progression via miR-204-3p/APC2 axis.8 However, the study about the function of circGDI2 in different cancers is very rare. This research certified that circGDI2 was distinctly downregulated in OSCC tissues and cells, and circGDI2 introduction overtly impeded proliferation, migration, invasion, and accelerated apoptosis in OSCC cells in vitro and hindered OSCC tumor in vivo growth, which was keeping with the previous report.9 Zhang et al also evidenced that upregulated exosomal circGDI2 decreased OSCC cell malignant behaviors via miR-424-5p/SCAI pathway.21 These data hinted that circGDI2 might be a possible target for OSCC treatment. Meanwhile, miR-454-3p was validated to be a target of circGDI2. Therefore, we then explored the association between circGDI2 and miR-454-3p in the following experiments.
The dysregulation of miRNAs is concerned with the carcinogenesis and pathogenesis of diversified malignancies, including OSCC.22–24 Overexpression of miR-146a accelerated cell proliferation and metastasis in OSCC25 and miR-4282 impeded tumor progression through LIN28B/ZBTB2 axis in OSCC cells and tumor growth in vivo.26 MiR-454-3p has been discovered to be a cancer-promoting factor in hepatocellular carcinoma27 and cervical cancer.15 Nevertheless, the precise mechanism of miR-454-3p in OSCC is blurry. In accordance with the result that has been reported,28 we attested that the level of miR-454-3p was incremental in OSCC tissues and cells. Simultaneously, the abundances of miR-454-3p and circGDI2 in OSCC tissues were reversely related. Furthermore, the effect of circGDI2 overexpression on OSCC cell progression was reversed by miR-454-3p restoration. The different expression patterns of miR-454-3p may be due to the different regulatory mechanisms and tissue specificity in a different type of cancers. Hence, it was speculated that circGDI2 might hinder OSCC development by reducing miR-454-3p expression.
FOXF2 is a transcription factor that is involved in the processes of tumorigenesis and is considered to be a biomarker for tumor diagnosis.17 For example, FOXF2 hindered Hela cell propagation and metastasis, and might serve as a prognostic biomarker for cervical cancer.29 FOXF2 was downregulated in HCC and might be a promising target in HCC remedy.30 MicroRNAs have been reported to function by repressing gene expression by sponging the 3ʹUTR of mRNAs.31 Moreover, FOXF2 expression was distinctly lessened in OSCC and silence of FOXF2 could dramatically accelerate the growth of OSCC cells by acting as a target of miR-96-5p.18 Consistent with this, our data certified that FOXF2 was downregulated in OSCC tissues and cells. Exhilaratingly, we found that miR-454-3p targeted FOXF2 and could regulate OSCC progression via sponging FOXF2. Then, we further certified that circGDI2 could regulate FOXF2 expression via miR-454-3p.
In conclusion, this study attested that circGDI2 was a tumor depressor and functioned by regulating FOXF2 expression via miR-454-3p in OSCC pathogenesis. This research might provide a potential target for OSCC treatment and illustrated the precise mechanism of circGDI2 in OSCC.
Abbreviations
OSCC, oral squamous cell carcinoma; FOXF2, forkhead box F2.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Thomson PJ. Perspectives on oral squamous cell carcinoma prevention-proliferation, position, progression and prediction. J Oral Pathol Med. 2018;47(9):803–807. doi:10.1111/jop.12733
2. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4–5):309–316. doi:10.1016/j.oraloncology.2008.06.002
3. Jiang S, Dong Y. Human papillomavirus and oral squamous cell carcinoma: A review of HPV-positive oral squamous cell carcinoma and possible strategies for future. Curr Probl Cancer. 2017;41(5):323–327. doi:10.1016/j.currproblcancer.2017.02.006
4. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi:10.15252/embj.2018100836
5. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-6
6. Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi:10.1016/j.cellsig.2017.10.009
7. Li B, Wang F, Li X, et al. Hsa_circ_0008309 may be a potential biomarker for oral squamous cell carcinoma. Dis Markers. 2018;2018:7496890. doi:10.1155/2018/7496890
8. Gao L, Zhao C, Li S, et al. circ-PKD2 inhibits carcinogenesis via the miR-204-3p/APC2 axis in oral squamous cell carcinoma. Mol Carcinog. 2019;58(10):1783–1794. doi:10.1002/mc.23065
9. Su W, Wang Y, Wang F, et al. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer. 2019;19(1):400. doi:10.1186/s12885-019-5593-5
10. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465. doi:10.1002/jcp.27486
11. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. doi:10.7150/thno.11543
12. Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2019;21(2):126–144. doi:10.1007/s12094-018-1910-8
13. Chitkara D, Mittal A, Mahato RI. miRNAs in pancreatic cancer: therapeutic potential, delivery challenges and strategies. Adv Drug Deliv Rev. 2015;81:34–52. doi:10.1016/j.addr.2014.09.006
14. Zuo J, Yu H, Xie P, et al. miR-454-3p exerts tumor-suppressive functions by down-regulation of NFATc2 in glioblastoma. Gene. 2019;710:233–239. doi:10.1016/j.gene.2019.06.008
15. Song Y, Guo Q, Gao S, Hua K. miR-454-3p promotes proliferation and induces apoptosis in human cervical cancer cells by targeting TRIM3. Biochem Biophys Res Commun. 2019;516(3):872–879. doi:10.1016/j.bbrc.2019.06.126
16. Guo JY, Wang YK, Lv B, Jin H. miR-454 performs tumor-promoting effects in oral squamous cell carcinoma via reducing NR3C2. J Oral Pathol Med. 2020;49(4):286–293. doi:10.1111/jop.13015
17. He W, Kang Y, Zhu W, et al. FOXF2 acts as a crucial molecule in tumours and embryonic development. Cell Death Dis. 2020;11(6):424. doi:10.1038/s41419-020-2604-z
18. Wang H, Ma N, Li W, Wang Z. MicroRNA-96-5p promotes proliferation, invasion and EMT of oral carcinoma cells by directly targeting FOXF2. Biol Open. 2020;9(3):bio049478. doi:10.1242/bio.049478
19. Gharat SA, Momin M, Bhavsar C. Oral squamous cell carcinoma: current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit Rev Ther Drug Carrier Syst. 2016;33(4):363–400. doi:10.1615/CritRevTherDrugCarrierSyst.2016016272
20. Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. doi:10.1038/onc.2017.89
21. Zhang Y, Tang K, Chen L, et al. Exosomal circGDI2 suppresses oral squamous cell carcinoma progression through the regulation of miR-424-5p/SCAI Axis. Cancer Manag Res. 2020;12:7501–7514. doi:10.2147/CMAR.S255687
22. Babaei K, Shams S, Keymoradzadeh A, et al. An insight of microRNAs performance in carcinogenesis and tumorigenesis; an overview of cancer therapy. Life Sci. 2020;240:117077. doi:10.1016/j.lfs.2019.117077
23. Harrandah AM, Mora RA, Chan EKL. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett. 2018;438:126–132. doi:10.1016/j.canlet.2018.09.019
24. Aali M, Mesgarzadeh AH, Najjary S, et al. Evaluating the role of microRNAs alterations in oral squamous cell carcinoma. Gene. 2020;757:144936. doi:10.1016/j.gene.2020.144936
25. Wang F, Ye LJ, Wang FJ, Liu HF, Wang XL. miR-146a promotes proliferation, invasion, and epithelial-to-mesenchymal transition in oral squamous carcinoma cells. Environ Toxicol. 2020;35(10):1050–1057. doi:10.1002/tox.22941
26. Zhang Y, Zhang Z, Huang W, Zeng J. MiR-4282 inhibits tumor progression through down-regulation of ZBTB2 by targeting LIN28B in oral squamous cell carcinoma. J Cell Physiol. 2020. doi:10.1002/jcp.29458
27. Li Y, Jiao Y, Fu Z, et al. High miR-454-3p expression predicts poor prognosis in hepatocellular carcinoma. Cancer Manag Res. 2019;11:2795–2802. doi:10.2147/CMAR.S196655
28. Guo JY, Wang YK, Lv B, Jin H. miR-454 performs tumor-promoting effects in oral squamous cell carcinoma via reducing NR3C2. J Oral Pathol Med. 2020;49(4):286–293. doi:10.1111/jop.13015
29. Zhang J, Zhang C, Sang L, et al. FOXF2 inhibits proliferation, migration, and invasion of Hela cells by regulating Wnt signaling pathway. Biosci Rep. 2018;38(5). doi:10.1042/BSR20180747
30. Shi Z, Liu J, Yu X, et al. Loss of FOXF2 expression predicts poor prognosis in hepatocellular carcinoma patients. Ann Surg Oncol. 2016;23(1):211–217. doi:10.1245/s10434-015-4515-2
31. Ni WJ, Leng XM. miRNA-dependent activation of mRNA translation. MicroRNA. 2016;5(2):83–86. doi:10.2174/2211536605666160825151201
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.