Back to Journals » Cancer Management and Research » Volume 12
CircFLNA Acts as a Sponge of miR-646 to Facilitate the Proliferation, Metastasis, Glycolysis, and Apoptosis Inhibition of Gastric Cancer by Targeting PFKFB2
Authors Qu J, Yang J, Chen M, Wei R, Tian J
Received 28 May 2020
Accepted for publication 8 August 2020
Published 7 September 2020 Volume 2020:12 Pages 8093—8103
DOI https://doi.org/10.2147/CMAR.S264674
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Juan Qu, 1 Jizhi Yang, 2 Ming Chen, 1 Rongna Wei, 1 Jingjing Tian 1
1Department of Gastroenterology, Tianjin Nankai Hospital, Tianjin, People’s Republic of China; 2Department of Traditional Chinese Medicine, Chen Tangzhuang Community Health Service Center, Hexi District, Tianjin, People’s Republic of China
Correspondence: Ming Chen Email [email protected]
Background: Many studies have confirmed that circular (circRNA) is involved in the development of gastric cancer (GC). However, the role of circFLNA in the progression of GC remains unclear.
Methods: Quantitative real-time PCR (qRT-PCR) was used to measure the relative expression of circFLNA, microRNA (miR)-646 and 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 2 (PFKFB2). Cell counting kit 8 (CCK8) assay, transwell assay and flow cytometry were performed to determine the proliferation, migration, invasion and apoptosis of cells, respectively. GC tumor xenograft models were built to confirm the function of circFLNA silencing on GC tumor growth in vivo. Furthermore, the lactate production, glucose consumption, ATP level and glucose uptake were detected to assess the glycolysis of cells. Then, the interaction between miR-646 and circFLNA or PFKFB2 was confirmed using dual-luciferase reporter assay. RNA immunoprecipitation (RIP) assay was used to verify the interaction between miR-646 and circFLNA further. In addition, Western blot (WB) analysis was employed to detect the relative protein expression of PFKFB2.
Results: Our results found that circFLNA was upregulated in GC tissues and cells. Silencing of circFLNA could suppress the proliferation, migration, invasion, glycolysis, and enhance the apoptosis of GC cells. Also, circFLNA knockdown reduced GC tumor volume and weight in vivo. Further experiments revealed that circFLNA could sponge miR-646, and miR-646 could target PFKFB2. The rescue experiments indicated that miR-646 inhibitor could reverse the suppressive effect of circFLNA silencing on GC progression, and PFKFB2 overexpression also could invert the inhibition effect of miR-646 on GC progression.
Conclusion: Our data concluded that circFLNA played a pro-cancer role in GC, which suggested that circFLNA might be a potential biomarker for GC treatment.
Keywords: GC, progression, circFLNA, miR-646, PFKFB2
Corrigendum for this paper has been published
Introduction
Gastric cancer (GC) is a malignant tumor that originates from gastric mucosa epithelium, and is a common disease that threatens human health.1,2 Early GC is generally not easy to detect, and most patients with GC have reached the advanced stage when diagnosed.3 More importantly, metastatic GC also affects the liver, kidney and respiratory function, which greatly reduces the prognosis of patients.4,5 In addition to proliferation and metastasis, glycolysis is also considered an important feature of cancer development.6,7 Therefore, elucidating the mechanisms affecting GC proliferation, metastasis and glycolysis are of great significance for the development of new therapeutic targets for GC.
Circular RNA (circRNA) is a covalently closed non-coding RNA produced by back-splicing.8 Functionally, it has been found that circRNA can indirectly regulate downstream gene expression by acting as a microRNA (miRNA) sponge.9,10 In recent years, some research has confirmed that circRNA exerts an essential regulatory function in the progress of diseases and has great potential to become a new type of clinical diagnostic marker, including cancer.11,12 In GC, circCCDC66 is considered as a potential biomarker for GC, and its high expression can accelerate GC proliferation and metastasis.13 Lu et al revealed that circ-CEP85L was underexpressed in GC, and it could sponge miR-942-5p to inhibit GC proliferation and invasion by targeting NFKBIA.14 Wang et al also showed that elevated circRBM33 expression could enhance the proliferation, metastasis and suppress the apoptosis of GC cells through regulating the miR-149/IL-6 pathway.15 Thus, circRNA is a vital biomarker for the development of new GC diagnosis and treatment targets.16,17
In order to identify the new circRNA, we analyzed the expression profile of circRNA in GC. By screening differentially expressed circRNAs in GC tumor tissues and adjacent normal tissues, we found that circFLNA was markedly upregulated in GC tissues. Nevertheless, the role of circFLNA in GC progression is unclear. Our research is to explore the function of circFLNA in the progression of GC and hope to provide new potential targets for GC treatment.
Materials and Methods
Samples Collection
45 pairs of GC tissues and adjacent normal tissues were acquired from 45 GC patients recruited from Tianjin Nankai Hospital. In addition, GC tissues were classified according to the different TNM stages (I–II and III–IV) of patients. All tissues were stored at −80°C, and informed consent was provided from all participants. Our study was approved by the Ethics Committee of Tianjin Nankai Hospital (IRB No.2017NK229).
Cell Culture and Transfection
Human GC cell lines (HGC27 and AGS) and normal gastric mucosal cell line (GES-1) were purchased from Biovector National Typical Culture Collection (Beijing, China). All cells were cultured in RPMI-1640 medium (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone) and 1% Penicillin/Streptomycin Solution (HyClone) at 37°C with 5% CO2.
Cell transfection was performed using Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA, USA) when the cells reached 60% confluences. Lentiviral short hairpin RNA against circFLNA and its control (sh-circFLNA#1/#2/#3 and sh-NC), pcDNA circFLNA and 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 2 (PFKFB2) overexpression plasmids (circFLNA and PFKFB2) or negative control (pcDNA) were obtained from Genechem (Shanghai, China). MiR-646 mimic and inhibitor (miR-646 and in-miR-646) or its controls (miR-NC and in-miR-NC) were bought from Ambion (Austin, TX, USA).
Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was obtained using PrimeScript RT Reagent Kit (Takara, Tokyo, Japan). Then, SYBR Green (for circFLNA; Takara) or TaqMan MicroRNA Assays (for miR-646; Applied Biosystems, Foster City, CA, USA) were used to perform qRT-PCR. β-actin or U6 was used as the inner control of circFLNA and PFKFB2 or miR-646, respectively. Relative expression was evaluated using the 2−ΔΔCt method. The detailed sequences were shown as follows: circFLNA, F 5ʹ-ACCACCATGACAACACCTACA-3ʹ, R 5ʹ-GCCAGCAGCTTTGGCATTTAC-3ʹ; miR-646, F 5ʹ-ACACTCCAGCTGGGAAGCAGCTGCCTC-3ʹ, R 5ʹ- CTCAACTGTGCTGCATTAGTTAGCTCAGA-3; PFKFB2, 5ʹ- AGTCCTACGACTTCTTTCGGC-3ʹ, R 5ʹ-TCTCCTCAGTGAGATACGCCT-3ʹ; β-actin, F 5ʹ-GTGCTATGTTGCTCTAGACTTC-3ʹ, R 5ʹ-ATGCCACAGGATTCCATACC-3ʹ; U6, F 5ʹ- AGCCCGCACTCAGAACATC-3ʹ, R 5ʹ-GCCACCAAGACAATCATCC-3ʹ.
Cell Counting Kit 8 (CCK8) Assay
HGC27 and AGS cells were sowed into 96-well plates (1 × 103 cells/well). At the indicated time points, 10 μL of CCK8 reagent (Dojindo, Kumamoto, Japan) was added into each well. After 2 h, the optical density (OD) value was recorded using a microplate reader at 450 nm.
Transwell Assay
Using the transwell chambers with or without Matrigel (BD Bioscience, San Diego, CA, USA), the invasion and migration of cells were determined, respectively. HGC27 and AGS cells mixed with serum-free medium were added into the upper chamber. The lower chamber was filled with complete medium. After maintained for 48 h, the upper chamber was removed, and the lower chamber cells were fixed with methanol and stained with crystal violet. Then, the cell number was counted under an inverted microscope (100 ×).
Flow Cytometry
Annexin V-FITC/PI Apoptosis Detection Kit was obtained from Yeasen (Shanghai, China). HGC27 and AGS cells were digested with trypsin and resuspended with binding buffer. Then, the cell suspension was added with Annexin V-FITC and PI staining solution at room temperature for 15 min away from light. The apoptosis of cells was measured using a flow cytometer.
Tumor Xenograft Models
AGS cells transfected with sh-NC or sh-circFLNA#3 were subcutaneously injected into the flank of BALB/c nude mice (4-week-old, male; Tengxin, Chongqing, China). Tumor length and width were measured with a vernier caliper every 7 days to calculate the tumor volume. After 28 days, the mice were euthanized and the tumors were collected for weighting. Animal experimental procedures were authorized by the Animal Ethics Committee of Tianjin Nankai Hospital, Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Measurement of Glucose Consumption, Lactate Production and ATP Level
The cultured medium was collected from transfected HGC27 and AGS cells. The Glucose Assay Kit, Lactate Assay Kit and ATP Assay Kit were purchased from Biovision (Milpitas, CA, USA). Glucose consumption, lactate production and ATP level were determined according to the manufacturer’s instructions, respectively. All values were normalized by protein content using BCA Kit (Beyotime, Shanghai, China).
Glucose Uptake Assay
Glucose Uptake Assay Kit was bought from Abcam (Cambridge, MA, USA). HGC27 and AGS cells were seeded in 96-well plates. After 12 h, the culture medium was removed and cells were washed with PBS. Then, the cells were incubated with 2-deoxyglucose (2-DG) for 20 min at room temperature. Then, the cells were washed with PBS to remove 2-DG. After the cells were lysed with extraction buffer, the cell lysates were freeze-thawed, heated, and incubated with the reaction mix. Cell fluorescence was detected by a microplate reader.
Dual-Luciferase Reporter Assay
The wild-type and mutant-type circFLNA and PFKFB2 3ʹUTR regions containing predicted and mutated binding sites of miR-646 were synthesized and sub-cloned into the psiCHECK-2 reporter vector (Promega, Madison, WI, USA), generating circFLNA WT/MUT and PFKFB2 3ʹUTR WT/MUT reporter vectors. These reporter vectors were transfected into HGC27 and AGS cells with miR-646 mimic and miR-NC using Lipofectamine 3000 Reagent. Dual-Luciferase Reporter Assay System (Promega) was employed to measure cell luciferase activity.
RNA Immunoprecipitation (RIP) Assay
EZ-Magna RIP Kit was purchased from Millipore (Billerica, MA, USA). HGC27 and AGS cells were transfected with miR-646 or miR-NC for 24 h. After that, the cells were lysed using RIP lysis buffer, and cell lysates were incubated with magnetic beads conjugated with antibodies against immunoglobulin G (RIP-IgG) or argonaute 2 (RIP-Ago2) overnight at 4°C. Then, the immunoprecipitated RNA was isolated using TRIzol reagent and circFLNA enrichment was assessed using qRT-PCR.
Western Blot (WB) Analysis
Proteins were extracted using RIPA buffer (Beyotime). After quantified using BCA Kit, the protein was separated by 10% SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were hatched with primary antibodies targeting PFKFB2 (1:4000, Abcam) or β-actin (1:10,000, Abcam), followed by reacting with secondary antibody (1:50,000, Abcam). Finally, the protein signal was detected using BeyoECL Moon (Beyotime).
Statistical Analysis
All data were presented as mean ± standard deviation and analyzed using GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical analysis was determined using Student’s t-test or one-way analysis of variance. The correlation analysis was conducted using Pearson correlation analysis. Statistically significant was defined as P < 0.05.
Results
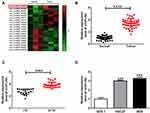
CircFLNA Was Upregulated in GC Tissues and Cells
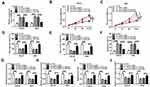
In GEO database (accession: GSE141977), we screened 20 differentially expressed circRNA (10 upregulated and 10 downregulated) in GC tumor tissues and adjacent normal tissues (Figure 1A). As one of the circRNA with a large expression difference, hsa_circRNA_105040 (circFLNA) was selected for this study. Through measuring the expression of circFLNA in GC tissues, we found that circFLNA was markedly upregulated in GC tissues compared to adjacent normal tissues (Figure 1B). Also, the expression of circFLNA in the tissues of GC patients with advanced-stage (III+IV) was significantly higher than that of GC patients with early-stage (I+II) (Figure 1C). In addition, Kaplan-Meier analysis showed that high circFLNA expression was significantly associated with the poor prognosis of GC patients (Supplementary Figure 1). Furthermore, we also found a high expression of circFLNA in GC cell lines (HGC27 and AGS) compared to normal gastric mucosal cell line (GES-1) (Figure 1D).
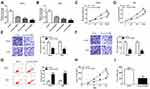
Interference of circFLNA Inhibited the Proliferation, Metastasis and Promoted the Apoptosis of GC Cells in vitro and Reduced the Tumor Growth of GC in vivo
In view of the high expression of circFLNA in GC, we silenced its expression to explore the role of circFLNA in GC using sh-circFLNA. As shown in Figure 2A and B, sh-circFLNA#1/#2/#3 could remarkably suppress the expression of circFLNA, especially sh-circFLNA#3 had the most obvious effect. Therefore, we used sh-circFLNA#3 for subsequent experiments. The results of CCK8 assay suggested that circFLNA knockdown could suppress the OD values of HGC27 and AGS cells, indicating that the proliferation of GC cells could be inhibited by circFLNA knockdown (Figure 2C and D). Besides, transwell assay was used to detect the migration and invasion of GC cells and the results showed that the migration and invasion rates of HGC27 and AGS cells also could be hindered by circFLNA silencing (Figure 2E and F). Moreover, we found that the apoptosis of HGC27 and AGS cells was remarkably promoted in the sh-circFLNA#3 group (Figure 2G). In addition, in constructed GC tumor xenograft models, we also discovered that knockdown of circFLNA could obviously reduce the tumor volume and weight of GC (Figure 2H–I).
CircFLNA Silencing Inhibited the Glycolysis of GC Cells
To explore the role of circFLNA on the glycolysis of GC cells, we measured the lactate production, glucose consumption and ATP level of GC cells. The results showed that circFLNA silencing could suppress the lactate production, glucose consumption and ATP level of HGC27 and AGS cells (Figure 3A–C). In addition, the glucose uptake of HGC27 and AGS cells also could be restrained by circFLNA knockdown (Figure 3D). These data suggested that circFLNA might promote the glycolysis of GC.
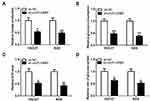
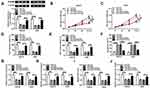
CircFLNA Could Sponge miR-646 in GC
To explore the mechanism of circFLNA in GC, a bioinformatic analysis was performed to identify putative targeted miRNAs for circFLNA. The circinteractome tool revealed that circFLNA could target the sequence of miR-646 (Figure 4A). The results of dual-luciferase reporter assay indicated that after transfected with miR-646 mimic, the luciferase activity of circFLNA WT was markedly reduced in HGC27 and AGS cells. However, miR-646 had not effect on the luciferase activity of circFLNA MUT (Figure 4B–C). Meanwhile, miR-646 also could remarkably increase the enrichment of circFLNA in RIP-Ago2 (Figure 4D). In GC tissues and cells, we found a significantly low miR-646 expression compared with that in adjacent normal tissues and GES-1 cells, respectively (Figure 4E–F). Also, correlation analysis revealed that there had a negative correlation between circFLNA and miR-646 (Figure 4G). To further confirm the regulation of circFLNA on miR-646, we transfected with pcDNA circFLNA overexpression plasmid into HGC27 and AGS cells. The markedly enhanced circFLNA expression suggested that transfection was successful (Figure 4H). Then, the detection results of miR-646 expression revealed that circFLNA overexpression could obviously inhibit miR-646 expression in HGC27 and AGS cells (Figure 4I). Therefore, our data suggested that circFLNA could directly interact with miR-646 in GC.
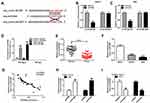
Inhibition of miR-646 Could Reverse the Regulation of circFLNA Silencing on GC Progression
For further confirming whether circFLNA regulated GC progression by sponging miR-646, sh-circFLNA#3 and in-miR-646 were co-transfected into HGC27 and AGS cells. Through measuring the expression of miR-646, we discovered that circFLNA knockdown could promote miR-646 expression, while the addition of in-miR-646 could reverse this effect, indicating that the transfection efficiency of both was excellent (Figure 5A). Subsequently, we determined the proliferation, metastasis, apoptosis and glycolysis of GC cells. CCK8 assay results were presented in Figure 5B and C, and it showed that miR-646 inhibitor could reverse the proliferation of HGC27 and AGS cells repressed by circFLNA silencing. Moreover, the inhibition effect of silenced circFLNA on the migration and invasion of HGC27 and AGS cells also were inverted by miR-646 inhibitor (Figure 5D–E). Compared with the control group, the decreased apoptosis rates in the sh-circFLNA#3 + in-miR-646 group suggested that miR-646 inhibitor could reverse the promoting effect of circFLNA knockdown on the apoptosis of HGC27 and AGS cells (Figure 5F). The original migration, invasion and apoptosis pictures for Figure 5D, E and F were shown in Supplementary Figure 2. In addition, the suppressive effect of circFLNA silencing on the lactate production, glucose consumption, ATP level and glucose uptake of HGC27 and AGS cells also could be reversed by miR-646 inhibitor (Figure 5G-J). These data revealed that circFLNA regulated the proliferation, metastasis, apoptosis and glycolysis of GC cells by sponging miR-646.
PFKFB2 Could Serve as a Target of miR-646 in GC
Using the Targetscan tool, we uncovered that PFKFB2 3ʹUTR had binding sites with miR-646 (Figure 6A). Then, we used dual-luciferase reporter assay to confirm the interaction between PFKFB2 and miR-646. It was found that the miR-646 overexpression could remarkably inhibit the luciferase activity of PFKFB2 3ʹUTR WT without affecting the luciferase activity of PFKFB2 3ʹUTR MUT (Figure 6B-C). Besides, we measured the expression of PFKFB2 in GC tissues and cells and discovered that PFKFB2 protein level was markedly enhanced compared to the corresponding negative controls (Figure 6D-E). At the same time, the results of correlation analysis indicated that PFKFB2 expression was negatively correlated with miR-646 expression and positively correlated with circFLNA expression in GC (Figure 6F-G). Furthermore, by detecting the expression of PFKFB2 using WB analysis, we discovered that miR-646 overexpression could suppress PFKFB2 expression (Figure 6H), and miR-646 inhibitor also could reverse the inhibition effect of circFLNA silencing on PFKFB2 expression (Figure 6I). All data suggested that PFKFB2 was a target of miR-646 and circFLNA regulated PFKFB2 expression by sponging miR-646.
PFKFB2 Overexpression Reversed the Suppression Effect of miR-646 on GC Progression
To determine that miR-646 regulated GC progression via targeting PFKFB2, we co-transfected with miR-646 mimic and pcDNA PFKFB2 overexpression plasmid into HGC27 and AGS cells. As shown in Figure 7A, the addition of pcDNA PFKFB2 overexpression plasmid relieved the inhibition effect of miR-646 on PFKFB2 expression, indicating that the transfection of them was effective. By detecting the proliferation, migration and invasion of HGC27 and AGS cells, we found that miR-646 overexpression could restrain the proliferation, migration and invasion of GC cells, while this effect could be reversed by PFKFB2 overexpression (Figure 7B-E). Meanwhile, miR-646 enhanced the apoptosis of HGC27 and AGS cells, and PFKFB2 overexpression also could reverse the promoting effect of miR-646 on GC cell apoptosis (Figure 7F). The original migration, invasion and apoptosis pictures for Figure 7D, E and F were presented in Supplementary Figure 2. Furthermore, the suppressive effects of miR-646 mimic on the lactate production, glucose consumption, ATP level and glucose uptake of HGC27 and AGS cells also were recovered by overexpressed PFKFB2 (Figure 7G-J). Hence, these results revealed that miR-646 regulated the progression of GC by targeting PFKFB2.
Discussion
There are many reports about the important role of circRNA in GC proliferation, metastasis and apoptosis.18,19 However, few studies have been conducted on circRNAs that affect the glycolysis of GC cells. Here, our study focused on a circRNA, circFLNA, which was significantly upregulated in GC, and explored its functionality in GC proliferation, metastasis, apoptosis and glycolysis. We discovered that circFLNA had elevated expression in GC tissues and cells. Loss-functional experiments indicated that silencing of circFLNA could restrain the proliferation, migration, invasion, and accelerate the apoptosis of GC cells. Further, animal experiments also revealed that circFLNA knockdown had an inhibitory regulation on GC tumor growth in vivo. In addition, we also found that the glycolysis of GC cells was decreased with the loss of circFLNA. Previous research proved that circFLNA was highly expressed in laryngeal squamous cell carcinoma (LSCC) and could regulate cell migration to promote LSCC progression.20 Therefore, similar to the previous results, our results also indicated that circFLNA played a positive role in GC progression.
MiR-646 is a widely downregulated miRNA in cancers.21 In many research, miR-646 is considered to be an anticancer factor that can be regulated by many cancer-promoting circRNAs. For example, circ_0000267 could sponge miR-646 to enhance the proliferation and metastasis of hepatocellular carcinoma.22 The recent study revealed that circ-CCND1 could accelerate the proliferation of LSCC via sponging miR-646.23 In GC, miR-646 had been shown to inhibit cell proliferation and metastasis by targeting FOXK1,24 and miR-646 could be targeted by circ_0081143 to participate in the regulation of GC cisplatin resistance.25 In our research, we suggested that circFLNA could interact with miR-646 in GC, and miR-646 was indeed lowly expressed in GC. Further rescue tests also confirmed that miR-646 was involved in the regulation of circFLNA on GC progression. So, our research showed that circFLNA could sponge miR-646 to exert its oncogenic role in GC.
PFKFB2 is a bifunctional isomeric enzyme and a regulatory molecule that controls glycolysis in eukaryotes.26,27 Due to the important role of glycolysis in cancer progression, the expression of PFKFB2 is associated with cancer progression.28,29 Our data showed that PFKFB2 was a target of miR-646, and its expression was positively regulated by circFLNA and negatively regulated by miR-646. In addition, the reversal effect of PFKFB2 on miR-646 regulation also further confirmed that miR-646 targeted PFKFB2 to hinder the proliferation, metastasis, glycolysis, and increase the apoptosis of GC. Our research discovered the presence of the circFLNA/miR-646/PFKFB2 axis, which revealed the potential mechanism by which circFLNA regulated GC progression.
In summary, we demonstrated that circFLNA was an upregulated circRNA in GC, which could enhance GC proliferation, metastasis, glycolysis, and apoptosis inhibition by regulating the miR-646/PFKFB2 axis. Our findings suggested that circFLNA might be an oncogene to regulate the development of GC, revealing that it might be a potential target for GC therapy.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Molina-Castro S, Pereira-Marques J, Figueiredo C, Machado JC, Varon C. Gastric cancer: basic aspects. Helicobacter. 2017;22(Suppl):1. doi:10.1111/hel.12412
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Digklia A, Wagner AD. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403–2414. doi:10.3748/wjg.v22.i8.2403
4. Lee SY, Oh SC. Changing strategies for target therapy in gastric cancer. World J Gastroenterol. 2016;22(3):1179–1189. doi:10.3748/wjg.v22.i3.1179
5. Pellino A, Riello E, Nappo F, et al. Targeted therapies in metastatic gastric cancer: current knowledge and future perspectives. World J Gastroenterol. 2019;25(38):5773–5788. doi:10.3748/wjg.v25.i38.5773
6. Yuan LW, Yamashita H, Seto Y. Glucose metabolism in gastric cancer: the cutting-edge. World J Gastroenterol. 2016;22(6):2046–2059. doi:10.3748/wjg.v22.i6.2046
7. Liu Y, Zhang Z, Wang J, et al. Metabolic reprogramming results in abnormal glycolysis in gastric cancer: a review. Onco Targets Ther. 2019;12:1195–1204. doi:10.2147/OTT.S189687
8. Li X, Yang L, Chen LL, Biogenesis T. Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
9. Tang Q, Hann SS. Biological Roles and Mechanisms of Circular RNA in Human Cancers. Onco Targets Ther. 2020;13:2067–2092. doi:10.2147/OTT.S233672
10. Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. doi:10.1007/978-981-13-1426-1_6
11. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
12. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-6
13. Xu G, Chen Y, Fu M, et al. Circular RNA CCDC66 promotes gastric cancer progression by regulating c-Myc and TGF-beta signaling pathways. J Cancer. 2020;11(10):2759–2768. doi:10.7150/jca.37718
14. Lu J, Wang YH, Huang XY, et al. circ-CEP85L suppresses the proliferation and invasion of gastric cancer by regulating NFKBIA expression via miR-942-5p. J Cell Physiol. 2020. doi:10.1002/jcp.29556
15. Wang N, Lu K, Qu H, et al. CircRBM33 regulates IL-6 to promote gastric cancer progression through targeting miR-149. Biomed Pharmacother. 2020;125:109876. doi:10.1016/j.biopha.2020.109876
16. Jiang F, Hong F, Shah MW, Shen X. Circular RNAs as diagnostic biomarkers in gastric cancer: A meta-analysis review. Pathol Res Pract. 2019;215(6):152419. doi:10.1016/j.prp.2019.04.011
17. Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi:10.7150/ijms.28047
18. Hu K, Qin X, Shao Y, Zhou Y, Ye G, Circular XS. RNA MTO1 suppresses tumorigenesis of gastric carcinoma by sponging miR-3200-5p and targeting PEBP1. Mol Cell Probes. 2020;52:101562. doi:10.1016/j.mcp.2020.101562
19. Ma Y, Cong X, Zhang Y, Yin X, Zhu Z, Xue Y. CircPIP5K1A facilitates gastric cancer progression via miR-376c-3p/ZNF146 axis. Cancer Cell Int. 2020;20(1):81. doi:10.1186/s12935-020-1122-5
20. Wang JX, Liu Y, Jia XJ, et al. Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA-miR-486-3p-FLNA axis. Cancer Cell Int. 2019;19(1):196. doi:10.1186/s12935-019-0924-9
21. Dai H, Hou K, Cai Z, Zhou Q, Zhu S. Low-level miR-646 in colorectal cancer inhibits cell proliferation and migration by targeting NOB1 expression.. Oncol Lett. 2017;14(6):6708–6714. doi:10.3892/ol.2017.7032
22. Pan H, Tang L, Jiang H, et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem. 2019;120(7):11350–11357. doi:10.1002/jcb.28411
23. Zang Y, Li J, Wan B, Tai Y. circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646. J Cell Mol Med. 2020;24(4):2423–2433. doi:10.1111/jcmm.14925
24. Zhang P, Tang WM, Zhang H, et al. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer. 2017;117(4):525–534. doi:10.1038/bjc.2017.181
25. Xue M, Li G, Fang X, Wang L, Jin Y, Zhou Q. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int. 2019;19(1):25. doi:10.1186/s12935-019-0737-x
26. Tang N, Zhang J, Fu X, Xie W, Qiu Y. PP2Acalpha inhibits PFKFB2-induced glycolysis to promote termination of liver regeneration. Biochem Biophys Res Commun. 2020. doi:10.1016/j.bbrc.2020.03.002
27. Moon JS, Jin WJ, Kwak JH, et al. Androgen stimulates glycolysis for de novo lipid synthesis by increasing the activities of hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 in prostate cancer cells. Biochem J. 2011;433(1):225–233. doi:10.1042/BJ20101104
28. He Z, You C, Zhao D. Long non-coding RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated stromal cells-mediated glycolysis and invasion of glioma cells. Biochem Biophys Res Commun. 2018;500(3):569–576. doi:10.1016/j.bbrc.2018.04.091
29. Zhao L, Ji G, Le X, et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017;77(6):1369–1382. doi:10.1158/0008-5472.CAN-16-1615
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.