Back to Journals » Cancer Management and Research » Volume 12
CircEPSTI1 Promotes the Progression of Non-Small Cell Lung Cancer Through miR-145/HMGB3 Axis
Received 7 March 2020
Accepted for publication 27 June 2020
Published 4 August 2020 Volume 2020:12 Pages 6827—6836
DOI https://doi.org/10.2147/CMAR.S252893
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Rudolph Navari
Yuanyuan Xie,1 Li Wang,2 Danfen Yang2
1Department of Geriatrics, Affiliated Hospital of Yan’an University, Yan’an 716000, Shaanxi, People’s Republic of China; 2Department of Respiratory Medicine, Affiliated Hospital of Yan’an University, Yan’an 716000, Shaanxi, People’s Republic of China
Correspondence: Danfen Yang Email [email protected]
Background: The high expression of circular RNA circEPSTI1 (hsa_circRNA_000479) has been reported to be associated with the malignant potential of ovarian cancer cells and triple-negative breast cancer cells. However, the expression profile and function of circEPSTI1 in non-small cell lung cancer (NSCLC) are not fully addressed.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was applied to measure the RNA expression of circEPSTI1, relevant microRNAs (miRNAs) and high mobility group box 3 (HMGB3) in NSCLC tissues and cells. Cell counting kit 8 (CCK8) assay, colony formation and transwell assays were conducted to detect the capacities of proliferation, colony formation and metastasis in NSCLC cells. Western blot assay was performed to detect the expression of metastasis-associated proteins and HMGB3. Animal experiment was carried out to confirm the function of circEPSTI1 in vivo. The combination between miR-145 and circEPSTI1 or HMGB3 was verified by dual-luciferase reporter assay, RNA pull-down and RIP assays.
Results: CircEPSTI1 was abnormally up-regulated in NSCLC tissues and cells in comparison with that in normal tissues and cells. The high expression of circEPSTI1 was associated with the low survival rate of NSCLC patients. CircEPSTI1 accelerated the proliferation, colony formation and motility of NSCLC cells in vitro. CircEPSTI1 silencing restrained the NSCLC tumor growth in vivo. miR-145 was validated as a target of circEPSTI1 in NSCLC cells. HMGB3 was a direct downstream target of miR-145 in NSCLC cells. The decreased abilities of proliferation, colony formation and metastasis caused by the silencing of circEPSTI1 were reversed by the depletion of miR-145 or the accumulation of HMGB3 in NSCLC cells.
Conclusion: CircEPSTI1 aggravated the progression of NSCLC through elevating the expression of HMGB3 via sponging miR-145.
Keywords: NSCLC, circEPSTI1, miR-145, HMGB3, proliferation, colony formation, metastasis
Introduction
Lung cancer is the leading cause of cancer-associated death globally, and more than 80% of lung cancer patients were identified with non-small cell lung cancer (NSCLC).1,2 The treatment strategies for NSCLC patients in clinic were chemotherapy, radiotherapy and targeted therapy. Nevertheless, the prognosis of NSCLC patients was poor owning to the recurrence and metastasis.3 Therefore, it is urgent to find crucial targets and effective therapeutic methods to improve the survival rate of NSCLC patients.
Circular RNAs (circRNAs) could modulate the biological processes through acting as microRNA (miRNA) sponges or competitive endogenous RNAs (ceRNAs).4–6 Wang et al claimed that circ_0001658 accelerated the growth and motility of osteosarcoma cells through miR-382-5p.7 Abulizi et al demonstrated that circ_0071662 restrained the progression of bladder cancer through miR-146b-3p.8 We concentrated on the role of circEPSTI1 (hsa_circRNA_000479) in NSCLC. Xie et al reported that circEPSTI1 promoted the malignant potential of ovarian cancer cells through miR-942.9 Hou and Chen claimed that circEPSTI1 facilitated the progression of triple-negative breast cancer.10 However, there are no reports focused on the biological role of circEPSTI1 in NSCLC.
miRNAs are a group of non-coding RNAs that are involved in the pathogenesis of cancers.11–13 MiRNAs could bind to messenger RNAs (mRNAs) to regulate their stability or impede their translation. MiR-145 has been reported to be down-regulated in NSCLC, and miR-145 inhibited the epithelial-mesenchymal transition (EMT) of NSCLC cells through ZEB2.14 Herein, we intended to search the potential mechanism of miR-145 in NSCLC.
High mobility group box 3 (HMGB3) has been confirmed as an oncogene in NSCLC, and its high expression was related to the poor prognosis of NSCLC patients.15 Besides, Song et al reported that HMGB3 accelerated the viability, colony formation and reduced the apoptosis of NSCLC cells.16 Nevertheless, the underlying network of HMGB3 in NSCLC remains poorly understood.
This study first assessed the biological role of circEPSTI1 in vivo and in vitro in NSCLC. Subsequently, the downstream RNA or protein was searched to illustrate the working mechanism by which circEPSTI1 promoting the progression of NSCLC.
Materials and Methods
Tissue Samples Collection
Sixty pairs of NSCLC specimens and corresponding normal specimens were obtained from NSCLC patients in Affiliated Hospital of Yan’an University. The tissues were pathologically identified by two experienced pathologists. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki, and that all subjects provided written informed consent and the approach was permitted by the Ethics Committee of Affiliated Hospital of Yan’an University.
Cell Culture
NSCLC cell lines, including A549, H1299, Calu-3, Calu-6 and normal human bronchial epithelial cell line HBE1 were all obtained from BeNa Culture Collection (Beijing, China). All cells were cultivated in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 units/mL penicillin and 100 μg/mL streptomycin in a 5% CO2 incubator at 37°C.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNAiso (Takara, Otsu, Japan) was utilized to extract total RNA from NSCLC tissues and cells. The complementary DNA (cDNA) was acquired using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The abundance of gene expression (2−ΔΔCt method) was normalized to U6 small nuclear RNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). SYBR-Green qPCR Mix (Bio-Rad, Hercules, CA, USA) was used in this study. The primers used in this study were as follows: circEPSTI1, Forward (F), AAGCTGAAGAAGCTGAACTC, Reverse (R), GTGTATGCACTTGTGTATTGC; miR-145, F, GCCGCGTCCAGTTTTCCCAGG, R, GTGCAGGGTCCGAGGT; miR-1248, F, ACCTTCTTGTATAAGCACTG, R, TTTAGCACAGTGCTTATACA; miR-1264, F, CAAGTCTTATTTGAGCACC, R, AACAGGTGCTCAAATAAGA; miR-370, F, GCCTGCTGGGGTGGAACCT, R, ACCAGGTTCCACCCCAGCA; miR-600, F, ACTTACAGACAAGAGCCTTG, R, GAGCAAGGCTCTTGTCTGTA; HMGB3, F, GACCAGCTAAGGGAGGCAA, R, ACAGGAAGAATCCAGACGGT; U6, F, CTCGCTTCGGCAGCACA, R, AACGCTTCACGAATTTGCGT; GAPDH, F, GGAGCGAGATCCCTCCAAAAT, R, GGCTGTTGTCATACTTCTCATGG.
Cell Transfection
Small hairpin RNA negative control (sh-NC), sh-circEPSTI1#1 (sh-circ#1), sh-circ#2, miR-NC, miR-145 mimics (miR-145), anti-NC, miR-145 inhibitor (anti-miR-145), vector and HMGB3 overexpression plasmid (HMGB3) were obtained from Genepharma (Shanghai, China). Transfection was conducted using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), and the transfection efficiency was evaluated by qRT-PCR.
Cell Counting Kit 8 (CCK8) Assay
The viability of NSCLC cells was evaluated by using Cell Counting Kit-8 (Beyotime, Shanghai, China). NSCLC cells were transferred into 96-well plates. 10 μL CCK8 reagent was mixed with the NSCLC cells after transfection for 0 h, 24 h, 48 h and 72 h for 3 h. The absorbance at 450 nm was detected to assess the proliferation rate of NSCLC cells.
Colony Formation Assay
After relevant treatment, A549 and H1299 cells were transferred to 6-well plates (200 cells/well) in triplicate. The culture medium was refreshed every 5 days. After incubation for 2 weeks, the colonies were fixed and then dyed with crystal violet (Sigma, St. Louis, MO, USA). The number of colonies in the control group and experimental group was counted and analyzed.
Transwell Assays
The motility of NSCLC cells was detected by transwell assays. In transwell invasion assay, the porous membrane was coated with matrigel (BD Biosciences, San Jose, CA, USA). A549 and H1299 cells were suspended in serum-free media, and 200 μL cell suspension was transferred to the pre-coated upper chambers. 500 μL culture medium added with 10% FBS was added to the lower chambers. After incubation for 24 h, the cells remained on the upper side of the membrane were removed mechanically. The invaded cells were stained with crystal violet. The cells in five random fields were counted using a microscope. The transwell migration assay was conducted using non-coated upper chambers according to the similar protocol.
Western Blot Assay
Transfected A549 and H1299 cells were collected and lysed using phosphate buffered saline (PBS) buffer and cell lysis buffer (Beyotime) added with protease inhibitor (Sigma), respectively. Equal amounts of protein (25 μg) from each group were separated on the separating gel, and the separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were then blocked for 1 h. The immunoblot was conducted using the following antibodies: anti-E-cadherin (anti-E-cad; ab1416, Abcam, Cambridge, MA, USA), anti-N-cadherin (anti-N-cad; ab18203, Abcam), anti-HMGB3 (AV35765, Sigma) and anti-GAPDH (ab37168, Abcam). After washed, the membranes were incubated with the secondary antibody (ab205718, Abcam). The protein bands were visualized using an enhanced chemiluminescence (ECL) system (Millipore).
Animal Study
The approach in this study was approved by the Institutional Animal Care and Use Committee at Affiliated Hospital of Yan’an University. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. A549 cells stably transfected with sh-circ#2 were inoculated into the right flank of the BALB/c nude mice (Orient Bio Inc, Seongnam, South Korea). The caliper was used to measure the volume of tumors every 7 d, and the mice bearing tumors were sacrificed after inoculation for 5 weeks. The expression of circEPSTI1 was detected using qRT-PCR assay.
Dual-Luciferase Reporter Assay
To test the relationship between miR-145 and circEPSTI1 in NSCLC cells, the sequences of circEPSTI1 (mutant or wild-type) were inserted into pSI-check2 vector (Promega, Madison, WI, USA), termed as circ-WT or circ-MUT, followed by co-transfection with miR-145 or miR-NC into A549 and H1299 cells. After 48 h transfection, dual-luciferase reporter assay system (Promega) was used to detect the luciferase activity. The relationship between miR-145 and HMGB3 was also verified according to the similar protocol.
RNA Pull-Down Assay
miR-145 was labeled with biotin, generating Bio-NC or Bio-miR-145. The A549 and H1299 lysate was incubated with Bio-NC or Bio-miR-145. QRT-PCR was utilized to determine the precipitated RNAs.
RNA Immunoprecipitation (RIP) Assay
A549 and H1299 cells were lysed using RIP lysis buffer (Haoran Biological Technology, Shanghai, China). Protein-A sepharose beads (Bio-Rad) were coated with anti-argonaute-2 (anti-ago2), and the anti-Immunoglobulin G (anti-IgG) was added instead of anti-ago2 as a control. QRT-PCR was conducted to detect the abundance of the immunoprecipitated RNA-RNA complexes.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD). Student’s t-test (for two groups) or one-way analysis of variance (ANOVA; for more than two groups) followed by Tukey’s test were used to analyze the differences. Survival curve of NSCLC patients was analyzed by Kaplan-Meier plot and Log rank test. P value less than 0.05 was considered statistical significance.
Results
CircEPSTI1 is Abnormally Up-Regulated in NSCLC Tissues and Cells
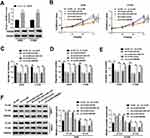
To explore the underlying involvement of circEPSTI1 in NSCLC, we conducted qRT-PCR to detect the expression of circEPSTI1 in NSCLC tissues and cells. As indicated in Figure 1A, the relative expression of circEPSTI1 was dramatically enhanced in NSCLC tissues in comparison with that in paired normal tissues. We also tested the abundance of circEPSTI1 in a panel of five cell lines, containing four NSCLC cell lines and one human bronchial epithelial cell line HBE1. The level of circEPSTI1 was higher in NSCLC cells than that in HBE1 cells (Figure 1B). Additionally, the high expression of circEPSTI1 was associated with the low survival rate of NSCLC patients (Figure 1C). In summary, circEPSTI1 was aberrantly up-regulated in NSCLC, and the expression of circEPSTI1 was negatively related to the prognosis of NSCLC patients.
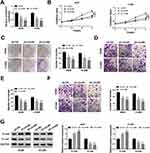
CircEPSTI1 Accelerates the Proliferation, Colony Formation and Metastasis of NSCLC Cells in vitro
We built A549 and H1299 cell lines stably transfected with sh-circEPSTI1 (sh-circ#1 or sh-circ#2) to assess the biological functions of circEPSTI1 in NSCLC. As shown in Figure 2A, the level of circEPSTI1 was significantly declined in sh-circ#1 and sh-circ#2 groups in contrast to that in sh-NC group. Besides, the proliferation was restrained with the depletion of circEPSTI1 in A549 and H1299 cells (Figure 2B). Moreover, the low expression of circEPSTI1 in NSCLC cells also caused a reduction in the number of colonies (Figure 2C). The migration and invasion capacities of A549 and H1299 cells were restrained with the silencing of circEPSTI1 (Figure 2D–F). Next, we found that the expression of E-cad was up-regulated, while the level of N-cad was reduced upon the intervention of circEPSTI1 in NSCLC cells (Figure 2G). Collectively, circEPSTI1 contributed to the malignant potential of NSCLC cells.
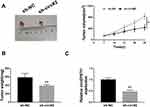
circEPSTI1 Promotes the Tumor Growth of NSCLC in vivo
We used murine xenograft model to evaluate whether circEPSTI1 exerted a same role in vivo. As exhibited in Figure 3A and B, the NSCLC tumors were smaller in mice injected with A549 cells stably transfected with sh-circ#2 than that in sh-NC group. The expression of circEPSTI1 was down-regulated in sh-circ#2 group in contrast to that in the control group (Figure 3C). These findings suggested that circEPSTI1 repressed the growth of NSCLC tumors in vivo.
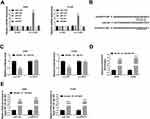
circEPSTI1 Acts as the Sponge of miR-145 in NSCLC Cells
There were five possible target miRNAs (miR-1248, miR-1264, miR-145, miR-370 and miR-600) of circEPSTI1 that predicted by Circinteractome database. Among these candidate target miRNAs, we found that the abundance of miR-145 was elevated with the interference of circEPSTI1 in NSCLC cells (Figure 4A). Hence, miR-145 was selected to verify its interaction with circEPSTI1. Dual-luciferase reporter assay was used to confirm the binding relationship between miR-145 and circEPSTI1 in NSCLC cells. The binding sequence between miR-145 and circEPSTI1 was shown in Figure 4B. We constructed luciferase reporter vector through inserting the sequence of circEPSTI1 containing the target sites of miR-145 into the luciferase vector, termed as circ-WT. And the mutant version named as circ-MUT. As mentioned in Figure 4C, a marked down-regulation of the luciferase activity was observed in miR-145 and circ-WT co-transfected group but not the miR-145 and circ-MUT co-transfected group, manifesting that miR-145 was a target of circEPSTI1 in A549 and H1299 cells. RNA pull-down assay exhibited that circEPSTI1 could be precipitated in Bio-miR-145 group in A549 and H1299 cells (Figure 4D). RIP assay showed that miR-145 and circEPSTI1 could be co-precipitated using ago2 antibody, suggesting that miR-145 and circEPSTI1 were existed in RNA induced silencing complex (RISC) (Figure 4E). Thus, these results validated miR-145 was a target gene of circEPSTI1 in NSCLC cells.
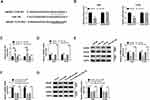
HMGB3 is a Target of miR-145 in NSCLC Cells
To illustrate how miR-145 functioned, Targetscan software was used to seek the downstream targets of miR-145. As shown in Supplementary Figure 1, among five candidate targets of miR-145 (PUM1, ADAM10, E2F6, ATM and HMGB3), ATM and HMGB3 were negatively regulated by miR-145, and the down-regulation of HMGB3 mRNA level was more obvious than ATM. Therefore, HMGB3 was chosen for further analysis. The complementary sequence between the 3ʹuntranslated region (3ʹUTR) of HMGB3 and miR-145 was shown in Figure 5A. The results of dual-luciferase reporter assay also confirmed the interaction between miR-145 and HMGB3 (Figure 5B). The level of miR-145 was prominently increased by the transfection of miR-145 mimics in NSCLC cells, while the transfection of anti-miR-145 revealed an inverse result (Figure 5C). We also tested the mRNA and protein expression of HMGB3 in NSCLC cells upon the overexpression or knockdown of miR-145. As mentioned in Figure 5D and E, the accumulation of miR-145 reduced the enrichment of HMGB3 mRNA and protein, while the mRNA and protein levels of HMGB3 were increased with the depletion of miR-145 in A549 and H1299 cells. In light of the above results, we further analyzed the regulatory modulation among circEPSTI1, miR-145 and HMGB3 in A549 and H1299 cells. The addition of anti-miR-145 recovered the mRNA and protein expression of HMGB3 in A549 and H1299 cells, which was down-regulated by the transfection of sh-circ#2 (Figure 5F and G). These data suggested that miR-145 could directly bind to HMGB3 in NSCLC cells.
The Knockdown of miR-145 or the Overexpression of HMGB3 Could Reverse the Suppressive Influences on the Proliferation, Colony Formation and Metastasis of NSCLC Cells Caused by circEPSTI1 Depletion
The abundance of HMGB3 protein was dramatically increased in A549 and H1299 cells transfected with HMGB3 overexpression plasmid (Figure 6A). A549 and H1299 cells were transfected with sh-NC, sh-circ#2, sh-circ#2 + anti-NC, sh-circ#2 + anti-miR-145, sh-circ#2 + vector or sh-circ#2 + HMGB3 to explore the functions of circEPSTI1, miR-145 and HMGB3 in NSCLC cells. The depletion of circEPSTI1 restrained the proliferation, colony formation and motility of NSCLC cells, while these suppressive influences were attenuated by the transfection of anti-miR-145 or HMGB3 (Figure 6B–F). In summary, circEPSTI1 contributed to the progression of NSCLC via miR-145/HMGB3 axis.
Discussion
Emerging researches have reported that circRNAs participated in the occurrence and development of cancers. Liang et al reported that circ_ABCB10 accelerated the development of breast cancer via miR-1271.17 Xu et al proved that circ_000984 facilitated the proliferation and motility of colon cancer cells via sponging miR-106b.18 Herein, we found that the expression of circEPSTI1 was higher in NSCLC tissues and cells, and it promoted the progression of NSCLC in vivo and in vitro. The oncogenic role of circEPSTI1 in NSCLC was alignment with its role in other cancers.9,19 MiR-145 served as a tumor suppressor in cancers, including NSCLC, gastric cancer, breast cancer and colorectal cancer.14,20–22 For example, Lei et al claimed that miR-145 played an anti-tumor role in gastric cancer through suppressing the metastasis of gastric cancer cells via MYO6.20 Ding et al found that miR-145 blocked the progression of breast cancer by restraining the proliferation and motility of breast cancer cells.21 Sheng et al claimed that miR-145 exerted a tumor suppressor role in colorectal cancer, and it suppressed the metastasis of colorectal cancer cells through PAK4-dependent pathway.22 Among five candidate miRNAs, miR-145 was negatively regulated by circEPSTI1 in NSCLC cells, and the putative target relationship between miR-145 and circEPSTI1 was validated by dual-luciferase reporter assay, RNA pull-down and RIP assays.
In addition, HMGB3 was found to be a target of miR-145 in NSCLC. HMG-Box family contains three members, named as HMGB1, HMGB2 and HMGB3.23–25 HMG-Box family could regulate DNA replication and transcription, etc. Besides, HMGB3 was an oncogene in diverse cancers. For instance, Song et al proved that HMGB3 acted as a target of miR-758 to promote the proliferation and metastasis of cervical cancer cells.26 Shi et al claimed that long non-coding RNA HOTTIP accelerated hypoxia-stimulated glycolysis via miR-615-3p/HMGB3 axis.27 As for NSCLC, Song et al proved that HMGB3 inhibition blocked the malignant potential of NSCLC cells, while HMGB3 silencing promoted the apoptosis of NSCLC cells.16 Shi et al reported that HOTTIP accelerated hypoxia-triggered glycolysis in NSCLC via miR-615-3p/HMGB3 axis.27 We found that the depletion of miR-145 or the overexpression of HMGB3 could alleviate the inhibitory effects of circEPSTI1 silencing on the progression of NSCLC. The pro-cancer role of HMGB3 was in agreement with the former reports.15,16,27
Taken together, we first elucidated the expression pattern and role of circEPST1 in NSCLC, and we found that circEPSTI1 promoted the proliferation, colony formation and metastasis of NSCLC cells through miR-145/HMGB3 axis.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Meza R, Meernik C, Jeon J, Cote ML. Lung cancer incidence trends by gender, race and histology in the United States, 1973–2010. PLoS One. 2015;10(3):e0121323. doi:10.1371/journal.pone.0121323
2. Riaz SP, Luchtenborg M, Coupland VH, Spicer J, Peake MD, Moller H. Trends in incidence of small cell lung cancer and all lung cancer. Lung Cancer. 2012;75(3):280–284. doi:10.1016/j.lungcan.2011.08.004
3. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi:10.1016/j.cell.2006.11.001
4. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. doi:10.1007/978-981-13-1426-1_6
5. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
6. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi:10.1093/bib/bbw053
7. Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem Funct. 2019. doi:10.1002/cbf.3452
8. Abulizi R, Li B, Zhang CG. Circ_0071662, a novel tumor biomarker, suppresses bladder cancer cell proliferation and invasion by sponging miR-146b-3p. Oncol Res. 2019. doi:10.3727/096504019x15740729375088
9. Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi:10.1111/jcmm.14260
10. Hou XX, Cheng H. Long non-coding RNA RMST silencing protects against middle cerebral artery occlusion (MCAO)-induced ischemic stroke. Biochem Biophys Res Commun. 2018;495(4):2602–2608. doi:10.1016/j.bbrc.2017.12.087
11. Leone P, Buonavoglia A, Fasano R, et al. Insights into the regulation of tumor angiogenesis by micro-RNAs. J Clin Med. 2019;8(12):2030. doi:10.3390/jcm8122030
12. Li J, Liang Y, Lv H, et al. miR-26a and miR-26b inhibit esophageal squamous cancer cell proliferation through suppression of c-MYC pathway. Gene. 2017;625:1–9. doi:10.1016/j.gene.2017.05.001
13. Mesci A, Huang X, Taeb S, et al. Targeting of CCBE1 by miR-330-3p in human breast cancer promotes metastasis. Br J Cancer. 2017;116(10):1350–1357. doi:10.1038/bjc.2017.105
14. Liu Q, Chen J, Wang B, et al. miR-145 modulates epithelial-mesenchymal transition and invasion by targeting ZEB2 in non-small cell lung cancer cell lines. J Cell Biochem. 2018. doi:10.1002/jcb.28126
15. Song N, Liu B, Wu JL, et al. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34(5):2599–2603. doi:10.1007/s13277-013-0807-y
16. Song N, Wang B, Feng G, et al. Knockdown of high mobility group box 3 impairs cell viability and colony formation but increases apoptosis in A549 human non-small cell lung cancer cells. Oncol Lett. 2019;17(3):2937–2945. doi:10.3892/ol.2019.9927
17. Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–1576.
18. Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683. doi:10.18632/oncotarget.21748
19. Chen D, Dixon BJ, Doycheva DM, et al. IRE1alpha inhibition decreased TXNIP/NLRP3 inflammasome activation through miR-17-5p after neonatal hypoxic-ischemic brain injury in rats. J Neuroinflammation. 2018;15(1):32. doi:10.1186/s12974-018-1077-9
20. Lei C, Du F, Sun L, et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017;8(10):e3101. doi:10.1038/cddis.2017.493
21. Ding Y, Zhang C, Zhang J, et al. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-beta1 expression. Int J Oncol. 2017;50(5):1701–1710. doi:10.3892/ijo.2017.3945
22. Sheng N, Tan G, You W, et al. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6(6):1331–1340. doi:10.1002/cam4.1029
23. Vaccari T, Beltrame M, Ferrari S, Bianchi ME. Hmg4, a new member of the Hmg1/2 gene family. Genomics. 1998;49(2):247–252. doi:10.1006/geno.1998.5214
24. Barreiro-Alonso A, Lamas-Maceiras M, Rodriguez-Belmonte E, Vizoso-Vazquez A, Quindos M, Cerdan ME. High mobility group B proteins, their partners, and other redox sensors in ovarian and prostate cancer. Oxid Med Cell Longev. 2016;2016:5845061. doi:10.1155/2016/5845061
25. Reeves R. High mobility group (HMG) proteins: modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair (Amst). 2015;36:122–136. doi:10.1016/j.dnarep.2015.09.015
26. Song T, Hou X, Lin B. MicroRNA-758 inhibits cervical cancer cell proliferation and metastasis by targeting HMGB3 through the WNT/beta-catenin signaling pathway. Oncol Lett. 2019;18(2):1786–1792. doi:10.3892/ol.2019.10470
27. Shi J, Wang H, Feng W, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol. 2019;862:172615. doi:10.1016/j.ejphar.2019.172615
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.